

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
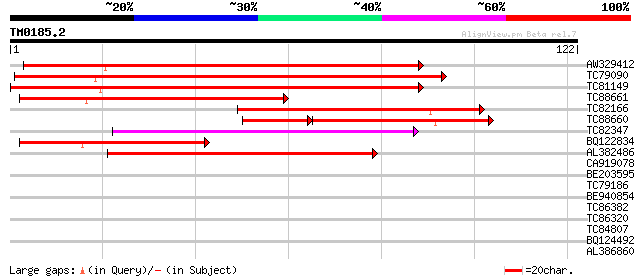
Query= TM0185.2
(122 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW329412 weakly similar to GP|11994591|db Ac transposase-like pr... 97 2e-21
TC79090 similar to PIR|T46111|T46111 probable transposase - Arab... 96 3e-21
TC81149 weakly similar to GP|11994591|dbj|BAB02646. Ac transposa... 96 4e-21
TC88661 weakly similar to GP|22507085|gb|AAM97760.1 putative tra... 65 8e-12
TC82166 similar to GP|10140707|gb|AAG13541.1 putative Tam3-trans... 50 2e-07
TC88660 PIR|T48044|T48044 hypothetical protein T12C14.220 - Arab... 42 3e-07
TC82347 46 4e-06
BQ122834 weakly similar to GP|20521440|dbj hypothetical protein~... 40 2e-04
AL382486 38 8e-04
CA919078 26 4.3
BE203595 26 4.3
TC79186 similar to GP|8777361|dbj|BAA96951.1 protein carboxyl me... 26 4.3
BE940854 similar to PIR|F96831|F96 hypothetical protein F18B13.1... 26 4.3
TC86382 similar to PIR|C84642|C84642 probable steroid binding pr... 25 5.6
TC86320 similar to GP|6957705|gb|AAF32449.1| unknown protein {Ar... 25 9.6
TC84807 similar to PIR|A96634|A96634 probable GCN4-complementing... 25 9.6
BQ124492 weakly similar to GP|9909168|dbj| putative transposable... 25 9.6
AL386860 similar to PIR|T02481|T024 probable protein kinase [imp... 25 9.6
>AW329412 weakly similar to GP|11994591|db Ac transposase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 569
Score = 97.1 bits (240), Expect = 2e-21
Identities = 45/87 (51%), Positives = 64/87 (72%), Gaps = 1/87 (1%)
Frame = +3
Query: 4 NELTKYLEDGLEERGS-LDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAGGR 62
++L KYLE+ + R S +ILNWWK++ RYPIL+ +AR++L P+ST+A E F+ GGR
Sbjct: 117 SDLDKYLEEPIFPRNSDFNILNWWKVHTPRYPILSMMARDVLGTPMSTLAPELAFNTGGR 296
Query: 63 VVDPYRSSLTPKTLEALICTQDWIKGK 89
++D RSSL P T EALICT DW++ +
Sbjct: 297 LLDSSRSSLNPDTREALICTHDWLRNE 377
>TC79090 similar to PIR|T46111|T46111 probable transposase - Arabidopsis
thaliana, partial (28%)
Length = 1068
Score = 96.3 bits (238), Expect = 3e-21
Identities = 45/94 (47%), Positives = 67/94 (70%), Gaps = 1/94 (1%)
Frame = +2
Query: 2 SKNELTKYLEDGLEER-GSLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAG 60
+K+EL +YLE+ L R D+L WWKLN +YP L+ +AR++L+IP+STV S+S F
Sbjct: 572 TKSELDQYLEESLLPRVPDFDVLGWWKLNKLKYPTLSKMARDILSIPVSTVPSDSIFDKK 751
Query: 61 GRVVDPYRSSLTPKTLEALICTQDWIKGKYSKSL 94
+ +D YRSSL P+T+EAL+C +DW++ +SL
Sbjct: 752 SKEMDQYRSSLRPETVEALVCAKDWMQYGAPESL 853
>TC81149 weakly similar to GP|11994591|dbj|BAB02646. Ac transposase-like
protein {Arabidopsis thaliana}, partial (11%)
Length = 806
Score = 95.5 bits (236), Expect = 4e-21
Identities = 45/90 (50%), Positives = 65/90 (72%), Gaps = 1/90 (1%)
Frame = +1
Query: 1 GSKNELTKYLEDGLEERG-SLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSA 59
G+K++L KYLE+ L R +ILNWWK++ RYP+L+ +AR +L IP+S VA E F+
Sbjct: 253 GAKSDLDKYLEEPLFPRNVDFNILNWWKVHTPRYPVLSMMARNVLGIPMSKVAPELAFNY 432
Query: 60 GGRVVDPYRSSLTPKTLEALICTQDWIKGK 89
GRV+D SSL P T++AL+C+QDWI+ +
Sbjct: 433 SGRVLDRDWSSLNPATVQALVCSQDWIRSE 522
>TC88661 weakly similar to GP|22507085|gb|AAM97760.1 putative transposase
{Oryza sativa (japonica cultivar-group)}, partial (4%)
Length = 521
Score = 64.7 bits (156), Expect = 8e-12
Identities = 31/59 (52%), Positives = 42/59 (70%), Gaps = 1/59 (1%)
Frame = +1
Query: 3 KNELTKYLEDGLE-ERGSLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAG 60
+N+L +YL D E + S DIL WWK N RYP+LA++ R++LA P+S+VASES S G
Sbjct: 334 QNDLERYLSDPPENDDPSFDILTWWKKNCVRYPVLATMVRDVLATPVSSVASESAXSTG 510
>TC82166 similar to GP|10140707|gb|AAG13541.1 putative Tam3-transposase
{Oryza sativa}, partial (6%)
Length = 574
Score = 50.4 bits (119), Expect = 2e-07
Identities = 30/56 (53%), Positives = 35/56 (61%), Gaps = 3/56 (5%)
Frame = +1
Query: 50 TVASESTFSAGGRVVDPYRSSLTPKTLEALICTQDWIKGK---YSKSLLSNEEVME 102
TVASES FS GR V P RS L TLEAL+C+Q+W + + YSK S E V E
Sbjct: 7 TVASESAFSTSGRDVTPQRSRLKEDTLEALMCSQNWFRTEMQGYSKIYASFECVDE 174
>TC88660 PIR|T48044|T48044 hypothetical protein T12C14.220 - Arabidopsis
thaliana, partial (2%)
Length = 865
Score = 42.0 bits (97), Expect(2) = 3e-07
Identities = 22/41 (53%), Positives = 28/41 (67%), Gaps = 2/41 (4%)
Frame = +3
Query: 66 PYRSSLTPKTLEALICTQDWIKGKY--SKSLLSNEEVMELE 104
PYRSSL+P+ EALICTQ+W+K + K L +EE LE
Sbjct: 51 PYRSSLSPEMAEALICTQNWLKPSFVDFKDLNLSEEYELLE 173
Score = 26.9 bits (58), Expect(2) = 3e-07
Identities = 11/15 (73%), Positives = 13/15 (86%)
Frame = +1
Query: 51 VASESTFSAGGRVVD 65
VASES FS GGR++D
Sbjct: 7 VASESAFSTGGRILD 51
>TC82347
Length = 750
Score = 45.8 bits (107), Expect = 4e-06
Identities = 20/66 (30%), Positives = 33/66 (49%)
Frame = +1
Query: 23 LNWWKLNASRYPILASIARELLAIPISTVASESTFSAGGRVVDPYRSSLTPKTLEALICT 82
+ WW + +P L+ +A ++ AIP E FS + R S+ +TLE C
Sbjct: 382 VQWWISHRKTFPTLSRLALDIFAIPAMATDCERAFSLAKLTLTSQRLSMKSQTLEEAQCL 561
Query: 83 QDWIKG 88
Q+W++G
Sbjct: 562 QNWLRG 579
>BQ122834 weakly similar to GP|20521440|dbj hypothetical protein~similar to
transposase {Oryza sativa (japonica cultivar-group)},
partial (10%)
Length = 553
Score = 40.4 bits (93), Expect = 2e-04
Identities = 18/42 (42%), Positives = 28/42 (65%), Gaps = 1/42 (2%)
Frame = +2
Query: 3 KNELTKYLEDGL-EERGSLDILNWWKLNASRYPILASIAREL 43
K+E+ +YLE+ L E DIL+WW+ N S+YP L+ A ++
Sbjct: 404 KSEMDEYLEEPLLSESKEFDILSWWRENRSKYPTLSRAASDI 529
>AL382486
Length = 492
Score = 38.1 bits (87), Expect = 8e-04
Identities = 19/58 (32%), Positives = 37/58 (63%)
Frame = +3
Query: 22 ILNWWKLNASRYPILASIARELLAIPISTVASESTFSAGGRVVDPYRSSLTPKTLEAL 79
I N+W ++ P+L++IA + + +PIS+ E +FS ++D R +L+ ++L+AL
Sbjct: 246 IENFWLGMVNQLPLLSNIALDYIWLPISSCTVERSFSIYNTILDDDRQNLSLESLKAL 419
>CA919078
Length = 515
Score = 25.8 bits (55), Expect = 4.3
Identities = 14/30 (46%), Positives = 20/30 (66%), Gaps = 3/30 (10%)
Frame = -3
Query: 76 LEALICTQDWIKGK---YSKSLLSNEEVME 102
LEAL+C+Q+W + + YSK + S E V E
Sbjct: 252 LEALMCSQNWFRTEMQGYSKIMASFECVDE 163
>BE203595
Length = 514
Score = 25.8 bits (55), Expect = 4.3
Identities = 10/16 (62%), Positives = 12/16 (74%)
Frame = +3
Query: 1 GSKNELTKYLEDGLEE 16
G NEL +YLEDG E+
Sbjct: 378 GHNNELVRYLEDGRED 425
>TC79186 similar to GP|8777361|dbj|BAA96951.1 protein carboxyl
methylase-like {Arabidopsis thaliana}, partial (71%)
Length = 911
Score = 25.8 bits (55), Expect = 4.3
Identities = 22/60 (36%), Positives = 27/60 (44%)
Frame = +2
Query: 48 ISTVASESTFSAGGRVVDPYRSSLTPKTLEALICTQDWIKGKYSKSLLSNEEVMELEKYE 107
I A + FS GG VVD SS K L C Q K SK + +EE + E+ E
Sbjct: 575 ILNAAMHAGFS-GGIVVDFPHSSKKRKEFLVLGCGQLSTKASLSKGKIEDEEKLSDEESE 751
>BE940854 similar to PIR|F96831|F96 hypothetical protein F18B13.11
[imported] - Arabidopsis thaliana, partial (1%)
Length = 368
Score = 25.8 bits (55), Expect = 4.3
Identities = 11/23 (47%), Positives = 14/23 (60%)
Frame = +2
Query: 75 TLEALICTQDWIKGKYSKSLLSN 97
TLEALIC +DW + +SN
Sbjct: 32 TLEALICAKDWFQHNSITKNVSN 100
>TC86382 similar to PIR|C84642|C84642 probable steroid binding protein
[imported] - Arabidopsis thaliana, complete
Length = 797
Score = 25.4 bits (54), Expect = 5.6
Identities = 18/46 (39%), Positives = 29/46 (62%), Gaps = 3/46 (6%)
Frame = +3
Query: 2 SKNE--LTKYLEDGLEERGSLDILNWWKLN-ASRYPILASIARELL 44
SKNE +T L DGL E+ + +LN W+ ++YP++A+I L+
Sbjct: 264 SKNEEDITSSL-DGLTEK-EIGVLNDWETKFVAKYPVVATIVS*LI 395
>TC86320 similar to GP|6957705|gb|AAF32449.1| unknown protein {Arabidopsis
thaliana}, partial (84%)
Length = 1440
Score = 24.6 bits (52), Expect = 9.6
Identities = 18/82 (21%), Positives = 35/82 (41%)
Frame = +3
Query: 4 NELTKYLEDGLEERGSLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAGGRV 63
N LT + E GL L + +W + N + + + + + +P++ +S ++ GR
Sbjct: 888 NILTSHAEIGLGFLLVLSLFSWQR-NIIQTFMYWQLLKLMYHVPVTAAYHQSVWAKIGRS 1064
Query: 64 VDPYRSSLTPKTLEALICTQDW 85
++P P L Q W
Sbjct: 1065INPLIHRHAPFLKTPLSAVQRW 1130
>TC84807 similar to PIR|A96634|A96634 probable GCN4-complementing protein
F23C21.2 [imported] - Arabidopsis thaliana, partial
(13%)
Length = 965
Score = 24.6 bits (52), Expect = 9.6
Identities = 17/75 (22%), Positives = 34/75 (44%), Gaps = 1/75 (1%)
Frame = -1
Query: 31 SRYPILASIARELLAIPISTV-ASESTFSAGGRVVDPYRSSLTPKTLEALICTQDWIKGK 89
S +P+L ++ ++ IP+ S S ++ V+ P +L+ A T D
Sbjct: 728 SNFPVLCTVLNSIICIPVPPYWRSTSRINSSSSVIAPISMALSSAVCPAASLT-DGFAPL 552
Query: 90 YSKSLLSNEEVMELE 104
SK+L + +E++
Sbjct: 551 LSKNLANGFFPLEMQ 507
>BQ124492 weakly similar to GP|9909168|dbj| putative transposable element
Tip100 protein {Oryza sativa (japonica cultivar-group)},
partial (8%)
Length = 694
Score = 24.6 bits (52), Expect = 9.6
Identities = 9/27 (33%), Positives = 18/27 (66%)
Frame = +1
Query: 33 YPILASIARELLAIPISTVASESTFSA 59
Y ++ + R ++ +P+ST +E +FSA
Sbjct: 136 YFLIDRLLRLIMTLPVSTATTERSFSA 216
>AL386860 similar to PIR|T02481|T024 probable protein kinase [imported] -
Arabidopsis thaliana, partial (7%)
Length = 382
Score = 24.6 bits (52), Expect = 9.6
Identities = 11/26 (42%), Positives = 15/26 (57%)
Frame = +1
Query: 82 TQDWIKGKYSKSLLSNEEVMELEKYE 107
T+DWI+ + S E ME E+YE
Sbjct: 283 TEDWIEVRAISSEQKEEATMENEEYE 360
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.139 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,698,864
Number of Sequences: 36976
Number of extensions: 42805
Number of successful extensions: 247
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 243
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 247
length of query: 122
length of database: 9,014,727
effective HSP length: 84
effective length of query: 38
effective length of database: 5,908,743
effective search space: 224532234
effective search space used: 224532234
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0185.2


