

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0182.6
(451 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
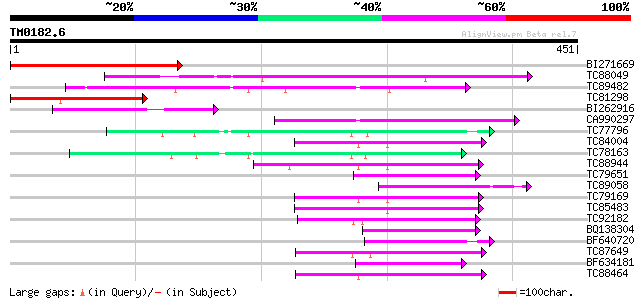
Score E
Sequences producing significant alignments: (bits) Value
BI271669 similar to GP|2668492|dbj metal-transporting P-type ATP... 237 6e-63
TC88049 similar to GP|13374852|emb|CAC34486. metal-transporting ... 235 2e-62
TC89482 similar to SP|Q9M3H5|AHM1_ARATH Potential cadmium/zinc-t... 144 9e-35
TC81298 similar to PIR|B96660|B96660 protein F2K11.18 [imported]... 112 3e-25
BI262916 similar to GP|15636781|gb copper-transporting P-type AT... 105 5e-23
CA990297 similar to GP|15636781|gb copper-transporting P-type AT... 102 3e-22
TC77796 H+-ATPase 73 2e-13
TC84004 type IIB calcium ATPase [Medicago truncatula] 69 5e-12
TC78163 H+-ATPase 67 1e-11
TC88944 type IIB calcium ATPase [Medicago truncatula] 65 4e-11
TC79651 similar to PIR|T52581|T52581 Ca2+-transporting ATPase (E... 63 2e-10
TC89058 similar to GP|19071218|gb|AAL84162.1 putative heavy meta... 63 3e-10
TC79169 type IIB calcium ATPase [Medicago truncatula] 62 5e-10
TC85483 type IIB calcium ATPase MCA5 [Medicago truncatula] 62 6e-10
TC92182 type IIA calcium ATPase [Medicago truncatula] 61 8e-10
BQ138304 homologue to GP|6688833|emb putative calcium P-type ATP... 59 4e-09
BF640720 homologue to PIR|S52728|S52 H+-transporting ATPase (EC ... 58 9e-09
TC87649 similar to SP|Q9SZR1|ACAA_ARATH Potential calcium-transp... 56 2e-08
BF634181 homologue to SP|Q08436|PMA3 Plasma membrane ATPase 3 (E... 55 6e-08
TC88464 similar to SP|Q9LU41|ACA9_ARATH Potential calcium-transp... 52 6e-07
>BI271669 similar to GP|2668492|dbj metal-transporting P-type ATPase
{Arabidopsis thaliana}, partial (13%)
Length = 509
Score = 237 bits (605), Expect = 6e-63
Identities = 115/137 (83%), Positives = 126/137 (91%)
Frame = +1
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQGSAVSLALQFACSVLVVACPCALGLA 60
+AGYFTYGVMA+SVTTFTFWS+FG I+P YQGS+VSLALQ ACSVLV+ACPCALGLA
Sbjct: 97 IAGYFTYGVMAISVTTFTFWSVFGPQIIPTAVYQGSSVSLALQLACSVLVIACPCALGLA 276
Query: 61 TPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIEN 120
TPTAVLVGTSLGAKRGLLLRGGNILEKFAMVN VVFDKTGTLT+G+PVVTK+V TCIEN
Sbjct: 277 TPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNTVVFDKTGTLTIGKPVVTKIVTGTCIEN 456
Query: 121 ANSSQTIENALSDVEIL 137
ANSSQT NALSD+E+L
Sbjct: 457 ANSSQTKINALSDIEVL 507
>TC88049 similar to GP|13374852|emb|CAC34486. metal-transporting ATPase-like
protein {Arabidopsis thaliana}, partial (38%)
Length = 1207
Score = 235 bits (600), Expect = 2e-62
Identities = 152/361 (42%), Positives = 212/361 (58%), Gaps = 20/361 (5%)
Frame = +1
Query: 76 GLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIENANSSQTIENALSDVE 135
GLL+RGG++LE+ A VN + DKTGTLT G+PVV+ + + E+ E
Sbjct: 1 GLLIRGGDVLERLAGVNYIALDKTGTLTRGKPVVSAIGSIHYGES--------------E 138
Query: 136 ILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTFLEEPGSGAVATIGNRKVYVGT 195
IL +AAAVE + HP+ KAI++ A+++ L G +E PG G +A I R V VG+
Sbjct: 139 ILHIAAAVEKTASHPIAKAIINKAESLE-LVLPPTKGQIVE-PGFGTLAEIDGRLVAVGS 312
Query: 196 LEWI----------------TRHGINNNILQEVECKNESFVYVG-VNDTLAGLIYFEDEV 238
LEW+ R +N++ +++ VYVG + + G I D V
Sbjct: 313 LEWVHERFNTRMNPSDLMNLERALMNHSSSTSSSKYSKTVVYVGREGEGIIGAIAISDIV 492
Query: 239 REDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVKPDQKKKFINELQ 298
REDA V L K+ I +LSGD+ A +A VGI D V + + P QK FI+ L+
Sbjct: 493 REDAESTVMRLKKKGIKTVLLSGDREEAVATIAETVGIENDFVKASLSPQQKSAFISSLK 672
Query: 299 K-DNIVAMVGDGINDAAALAASHIGIALGGGV--GAASEVSSIILMRDHLSQLLDALELS 355
+ VAMVGDGINDA +LAA+ +GIAL AAS+ +SIIL+ + +SQ++DAL+L+
Sbjct: 673 AAGHHVAMVGDGINDAPSLAAADVGIALQNEAQENAASDAASIILLGNKISQVIDALDLA 852
Query: 356 RLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTPSIAGALMGLSSIGVMTNSLLL 415
+ TM V QNL WA YN++ IPIAAGVL P +TPS++G LM +SSI V +NSLLL
Sbjct: 853 QATMAKVYQNLSWAVAYNVIAIPIAAGVLLPQFDFAMTPSLSGGLMAMSSILVGSNSLLL 1032
Query: 416 R 416
+
Sbjct: 1033K 1035
>TC89482 similar to SP|Q9M3H5|AHM1_ARATH Potential cadmium/zinc-transporting
ATPase HMA1 (EC 3.6.3.3) (EC 3.6.3.5). [Mouse-ear
cress], partial (41%)
Length = 1100
Score = 144 bits (362), Expect = 9e-35
Identities = 110/346 (31%), Positives = 178/346 (50%), Gaps = 24/346 (6%)
Frame = +3
Query: 45 ACSVLVVACPCALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTV 104
A ++V A PCAL +A P A S AK+G+LL+GG++L+ A + + FDKTGTLT
Sbjct: 15 ALGLMVAASPCALAVA-PLAYATAISSCAKKGILLKGGHVLDALASCHTIAFDKTGTLTT 191
Query: 105 GRPVVTKVVASTCIENANSSQTIENA---LSDVEILRLAAAVESNSVHPVGKAIVDAAQA 161
G V + N I + + E L +AAA+E + HP+G+A+V+ ++
Sbjct: 192 GGLVFKAIEPVYGHHIRNKESNISSCCVPTCEKEALAVAAAMEKGTTHPIGRAVVEHSEG 371
Query: 162 VNCLDAKVVDGTFLEEPGSGAVATIGN----------RKVYVGTLEWITRHGINNNILQE 211
N V + + PG G AT+ + K +G++++IT + + ++
Sbjct: 372 KNLPSVSVENFEYF--PGRGLTATVNSIESGAGGANLLKASLGSIDFITSFCQSEDESKK 545
Query: 212 V-ECKNES-----FVYVG-VNDTLAGLIYFEDEVREDARHVVDTLSKQ-DISVYMLSGDK 263
V E N S FV+ + + LI+ ED R V+ L + V ML+GD
Sbjct: 546 VKEAINASSYGSEFVHAALIINKKVTLIHLEDRPRPGVFDVIQELQDEAKFRVMMLTGDH 725
Query: 264 RNAAEHVASLVGIPKDKVLSGVKPDQKKKFINELQKD--NIVAMVGDGINDAAALAASHI 321
+A VAS VGI + +KP+ K + + ++ +D + MVG+GINDA ALAA+ +
Sbjct: 726 EYSARRVASAVGI--KEFHCNLKPEDKLRHVKDISRDMGGGLIMVGEGINDAPALAAATV 899
Query: 322 GIALGGGVGA-ASEVSSIILMRDHLSQLLDALELSRLTMTTVKQNL 366
GI L A A V+ ++L+R+++S + + SR T + +KQN+
Sbjct: 900 GIVLAHRASATAIAVADVLLLRENISAVPFCIAKSRQTTSLIKQNV 1037
>TC81298 similar to PIR|B96660|B96660 protein F2K11.18 [imported] -
Arabidopsis thaliana, partial (23%)
Length = 848
Score = 112 bits (280), Expect = 3e-25
Identities = 59/112 (52%), Positives = 79/112 (69%), Gaps = 3/112 (2%)
Frame = +1
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGT-HILPATAYQGSAVS--LALQFACSVLVVACPCAL 57
++ YF V+ +S++T+ W + G H P + S S LALQF SV+V+ACPCAL
Sbjct: 460 ISKYFVPIVIVLSLSTWISWFVAGKLHSYPKSWIPSSMNSFELALQFGISVMVIACPCAL 639
Query: 58 GLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVV 109
GLATPTAV+VGT +GA +G+L++GG LE VN +VFDKTGTLT+G+PVV
Sbjct: 640 GLATPTAVMVGTGVGATQGVLIKGGQALESAHKVNCIVFDKTGTLTIGKPVV 795
>BI262916 similar to GP|15636781|gb copper-transporting P-type ATPase
{Brassica napus}, partial (15%)
Length = 527
Score = 105 bits (261), Expect = 5e-23
Identities = 58/132 (43%), Positives = 79/132 (58%)
Frame = +3
Query: 35 GSAVSLALQFACSVLVVACPCALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAV 94
G+ AL F+ SV+V+ACPCALGLATPTAV+V T +GA G+L++GG LE+ MV V
Sbjct: 138 GNHFVFALMFSISVVVIACPCALGLATPTAVMVATGVGANNGVLIKGGESLERAQMVKYV 317
Query: 95 VFDKTGTLTVGRPVVTKVVASTCIENANSSQTIENALSDVEILRLAAAVESNSVHPVGKA 154
+FDKTGTLT G+ V S + + E L+L A+ E+ S HP+ KA
Sbjct: 318 IFDKTGTLTQGKATV-------------SVAKVFAGMDRGEFLKLVASAEAXSEHPLAKA 458
Query: 155 IVDAAQAVNCLD 166
I+ A+ + D
Sbjct: 459 ILQYARHFHFFD 494
>CA990297 similar to GP|15636781|gb copper-transporting P-type ATPase
{Brassica napus}, partial (19%)
Length = 591
Score = 102 bits (254), Expect = 3e-22
Identities = 68/196 (34%), Positives = 106/196 (53%), Gaps = 1/196 (0%)
Frame = +3
Query: 211 EVECKNESFVYVGVNDTLAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHV 270
E+E ++ + V N L G++ D ++ +A VV+ L K I+ M++GD A V
Sbjct: 9 ELEESAKTGILVACNGILIGVLGVADSLKREASVVVEGLQKMGITPVMVTGDNWRTARAV 188
Query: 271 ASLVGIPKDKVLSGVKPDQKKKFINELQKD-NIVAMVGDGINDAAALAASHIGIALGGGV 329
A VGI V + V P K + QKD +IVAMVGDGIND+ ALAA+ +G+A+G G
Sbjct: 189 AKEVGI--QDVRAEVMPAGKADVVRSFQKDGSIVAMVGDGINDSPALAAADVGMAIGAGT 362
Query: 330 GAASEVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNG 389
A E ++ +LMRD+L ++ A++LS+ ++ + ++ + G F G
Sbjct: 363 DIAIEAANYVLMRDNLEDVITAIDLSKKHFLVFD*IMYSPWPTILLPYRLLPGFYFLHWG 542
Query: 390 TMLTPSIAGALMGLSS 405
+ +AGA M LSS
Sbjct: 543 SSFHHGVAGACMALSS 590
>TC77796 H+-ATPase
Length = 3416
Score = 73.2 bits (178), Expect = 2e-13
Identities = 89/360 (24%), Positives = 141/360 (38%), Gaps = 52/360 (14%)
Frame = +1
Query: 78 LLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIEN----------ANSSQTI 127
L R +L K+ ++ + DKTGTLT+ + V K + + A +S+T
Sbjct: 1027 LQRE*QLLRKWQGMDVLCSDKTGTLTLNKLSVEKNLIEVFAKGVEKDYVILLAARASRTE 1206
Query: 128 ENALSDVEILRLAAAVES-----NSVHPVGKAIVDAAQAVNCLDAKVVDGTFLEEPGSGA 182
D I+ + A + VH VD A+ +DA DG + GA
Sbjct: 1207 NQDAIDAAIVGMLADPKEARAGVREVHFFPFNPVDKRTALTYIDA---DGNW-HRSSKGA 1374
Query: 183 VATIGN--------RKVYVGTLEWITRHGINNNILQEVECKNESFVYVGVNDTLAGLIYF 234
I N RK T++ G+ + + E + G GL+
Sbjct: 1375 PEQILNLCNCKEDVRKKAHSTIDKFAERGLRSLGVARQEIPEKDKDSPGAPWQFVGLLPL 1554
Query: 235 EDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHV-------------ASLVGIPKDKV 281
D R D+ + ++V M++GD+ A+ +SL+G KD
Sbjct: 1555 FDPPRHDSAETITRALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSSSLLGQSKDAA 1734
Query: 282 LS---------------GVKPDQKKKFINELQ-KDNIVAMVGDGINDAAALAASHIGIAL 325
+S GV P+ K + + +LQ + +I M GDG+NDA AL + IGIA+
Sbjct: 1735 VSALPVDELIEKADGFAGVFPEHKYEIVKKLQERKHICGMTGDGVNDAPALKRADIGIAV 1914
Query: 326 GGGVGAASEVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGIPIAAGVLF 385
AA S I+L LS ++ A+ SR +K Y I + I ++F
Sbjct: 1915 ADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKN-------YTIYAVSITIRIVF 2073
>TC84004 type IIB calcium ATPase [Medicago truncatula]
Length = 3655
Score = 68.6 bits (166), Expect = 5e-12
Identities = 48/181 (26%), Positives = 84/181 (45%), Gaps = 28/181 (15%)
Frame = +1
Query: 227 TLAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGI---------- 276
TL L+ +D VR + V T I+V M++GD N A+ +A GI
Sbjct: 2176 TLIALVGIKDPVRPGVKEAVQTCIAAGITVRMVTGDNINTAKAIAKECGILTDDGVAIEG 2355
Query: 277 ---------------PKDKVLSGVKPDQKKKFINELQK--DNIVAMVGDGINDAAALAAS 319
P+ +V++ P K K + L+ +VA+ GDG NDA AL +
Sbjct: 2356 PSFRELSDEQMKDIIPRIQVMARSLPLDKHKLVTNLRNMFGEVVAVTGDGTNDAPALHEA 2535
Query: 320 HIGIALG-GGVGAASEVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGIP 378
IG+A+G G A E + +I+M D+ + +++ ++ R +++ + + N+V +
Sbjct: 2536 DIGLAMGIAGTEVAKEKADVIIMDDNFATIVNVVKWGRAVYINIQKFVQFQLTVNVVALI 2715
Query: 379 I 379
I
Sbjct: 2716 I 2718
Score = 35.0 bits (79), Expect = 0.059
Identities = 29/112 (25%), Positives = 50/112 (43%), Gaps = 3/112 (2%)
Frame = +1
Query: 45 ACSVLVVACPCALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTV 104
A +++VVA P L LA ++ L+R + E + + DKTGTLT
Sbjct: 1435 AVTIIVVAVPEGLPLAVTLSLAFAMKKLMNDMALVRHLSACETMGSASCICTDKTGTLTT 1614
Query: 105 GRPVVTKV---VASTCIENANSSQTIENALSDVEILRLAAAVESNSVHPVGK 153
VV K+ +T ++ S+ ++ +S+ + L A+ N+ V K
Sbjct: 1615 NHMVVNKIWICENTTQLKGDESADELKTNISEGVLSILLQAIFQNTSAEVVK 1770
>TC78163 H+-ATPase
Length = 3295
Score = 67.4 bits (163), Expect = 1e-11
Identities = 81/368 (22%), Positives = 146/368 (39%), Gaps = 52/368 (14%)
Frame = +1
Query: 48 VLVVACPCALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRP 107
+L+ P A+ + +G+ +++G + + +E+ A ++ + DKTGTLT+ +
Sbjct: 994 LLIGGIPIAMPTVLSVTMAIGSHKLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKL 1173
Query: 108 VVTKVVASTCIENANSSQTI-----------ENALSDVEILRLAAAVESNS----VHPVG 152
V K + + + + ++A+ + LA E+ + VH +
Sbjct: 1174 TVDKDMIEVFAKGVDKDLVVLMAARASRLENQDAIDCAIVSMLADPKEARTGIKEVHFLP 1353
Query: 153 KAIVDAAQAVNCLDAKVVDGTFLEEPGSGAVATIGN---------RKVYVGTLEWITRHG 203
D A+ +DA + GA I N +KV+ ++ G
Sbjct: 1354 FNPTDKRTALTYIDA----AGNMHRVSKGAPEQILNLARNKAEIAQKVH-SMIDKFAERG 1518
Query: 204 INNNILQEVECKNESFVYVGVNDTLAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDK 263
+ + + E S G L+ D R D+ + +SV M++GD+
Sbjct: 1519 LRSLGVARQEVPEGSKDSPGGPWEFVALLPLFDPPRHDSAETIRRALDLGVSVKMITGDQ 1698
Query: 264 RNAAEHV-------------ASLVGIPKDKV--------------LSGVKPDQKKKFINE 296
+ +SL+G KD++ +GV P+ K + +
Sbjct: 1699 LAIGKETGRRLGMGTNMYPSSSLLGDNKDQLGAVSIDDLIENADGFAGVFPEHKYEIVKR 1878
Query: 297 LQ-KDNIVAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILMRDHLSQLLDALELS 355
LQ + +I M GDG+NDA AL + IGIA+ AA S I+L LS ++ A+ S
Sbjct: 1879 LQARKHICGMTGDGVNDAPALKIADIGIAVADSTDAARSASDIVLTEPGLSVIISAVLTS 2058
Query: 356 RLTMTTVK 363
R +K
Sbjct: 2059 RAIFQRMK 2082
>TC88944 type IIB calcium ATPase [Medicago truncatula]
Length = 1883
Score = 65.5 bits (158), Expect = 4e-11
Identities = 53/213 (24%), Positives = 93/213 (42%), Gaps = 30/213 (14%)
Frame = +3
Query: 195 TLEWITRHGINNNILQEVECKNESFVY--VGVND-TLAGLIYFEDEVREDARHVVDTLSK 251
T+E + L ++ +E V + VN T G++ +D VR R V
Sbjct: 606 TIEKFANEALRTLCLAYIDIHDEFLVGSPIPVNGYTCVGIVGIKDPVRPGVRESVAICRS 785
Query: 252 QDISVYMLSGDKRNAAEHVASLVGI------------------------PKDKVLSGVKP 287
I+V M++GD N A+ +A GI PK +V++ P
Sbjct: 786 AGITVRMVTGDNINTAKAIARECGILTDGIAIEGPEFREMSEKELLDIIPKIQVMARSSP 965
Query: 288 DQKKKFINELQK--DNIVAMVGDGINDAAALAASHIGIALG-GGVGAASEVSSIILMRDH 344
K + L+ + +VA+ GDG NDA AL + IG+A+G G A E + +I++ D+
Sbjct: 966 MDKHTLVKHLRTTFEEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDN 1145
Query: 345 LSQLLDALELSRLTMTTVKQNLWWAFIYNIVGI 377
S ++ + R +++ + + N+V +
Sbjct: 1146FSTIVTVAKWGRSVYINIQKFVQFQLTVNVVAL 1244
Score = 30.0 bits (66), Expect = 1.9
Identities = 19/60 (31%), Positives = 25/60 (41%)
Frame = +3
Query: 86 EKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIENANSSQTIENALSDVEILRLAAAVES 145
E + DKTGTLT V K I+ NSS + SD+ +A +ES
Sbjct: 96 ETMGSATTICSDKTGTLTTNHMTVVKACICGKIKEVNSSIDSSDFSSDLPDSAIAILLES 275
>TC79651 similar to PIR|T52581|T52581 Ca2+-transporting ATPase (EC 3.6.1.38)
ECA3 [imported] - Arabidopsis thaliana, partial (38%)
Length = 1546
Score = 63.2 bits (152), Expect = 2e-10
Identities = 33/102 (32%), Positives = 54/102 (52%), Gaps = 1/102 (0%)
Frame = +3
Query: 274 VGIPKDKVLSGVKPDQKKKFINELQKDN-IVAMVGDGINDAAALAASHIGIALGGGVGAA 332
+ + + + + V+P K+ + LQ N +VAM GDG+NDA AL + IGIA+G G A
Sbjct: 144 IALQRMALFTRVEPSHKRMLVEALQHQNEVVAMTGDGVNDAPALKKADIGIAMGSGTAVA 323
Query: 333 SEVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNI 374
S ++L D+ + ++ A+ R KQ + + NI
Sbjct: 324 KSASDMVLADDNFASIVAAVAEGRAIYNNTKQFIRYMISSNI 449
>TC89058 similar to GP|19071218|gb|AAL84162.1 putative heavy metal
transporter {Arabidopsis thaliana}, partial (10%)
Length = 1618
Score = 62.8 bits (151), Expect = 3e-10
Identities = 44/124 (35%), Positives = 70/124 (55%), Gaps = 2/124 (1%)
Frame = +1
Query: 294 INELQKDNIVAMVGDGINDAAALAASHIGIALG-GGVGAASEVSSIILMRDHLSQLLDAL 352
I E +KD AM+GDG+NDA ALA++ IGI++G G ASE IILM + L ++ +A+
Sbjct: 1 ITEFKKDGPTAMLGDGLNDAPALASADIGISMGISGSALASETGDIILMSNDLRKIPEAI 180
Query: 353 ELSRLTMTTVKQNLWWAFIYNIVGIPIA-AGVLFPVNGTMLTPSIAGALMGLSSIGVMTN 411
+L+R V +N+ + I + + +A AG P+ + + L+ V+ N
Sbjct: 181 KLARKARRKVIENIVLSVITKVAILALAIAG--HPIVWAAVLADVGTCLL------VILN 336
Query: 412 SLLL 415
S+LL
Sbjct: 337 SMLL 348
>TC79169 type IIB calcium ATPase [Medicago truncatula]
Length = 3565
Score = 62.0 bits (149), Expect = 5e-10
Identities = 46/179 (25%), Positives = 79/179 (43%), Gaps = 28/179 (15%)
Frame = +2
Query: 227 TLAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGI---------- 276
TL ++ +D VR + V ISV M++GD N A+ +A GI
Sbjct: 2102 TLITIVGIKDPVRPGVKEAVQKCLAAGISVRMVTGDNINTAKAIAKECGILTEGGVAIEG 2281
Query: 277 ---------------PKDKVLSGVKPDQKKKFINELQK--DNIVAMVGDGINDAAALAAS 319
P+ +V++ P K + L+ +VA+ GDG NDA AL S
Sbjct: 2282 PEFRNLSEEQMKDIIPRIQVMARSLPLDKHTLVTRLRNMFGEVVAVTGDGTNDAPALHES 2461
Query: 320 HIGIALG-GGVGAASEVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGI 377
IG+A+G G A E + +I+M D+ + ++ + R +++ + + N+V +
Sbjct: 2462 DIGLAMGIAGTEVAKENADVIIMDDNFTTIVKVAKWGRAIYINIQKFVQFQLTVNVVAL 2638
Score = 37.7 bits (86), Expect = 0.009
Identities = 78/356 (21%), Positives = 138/356 (37%), Gaps = 33/356 (9%)
Frame = +2
Query: 45 ACSVLVVACPCALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTV 104
A +++VVA P L LA ++ L+R + E + + DKTGTLT
Sbjct: 1367 AVTIIVVAIPEGLPLAVTLSLAFAMKKLMNDRALVRHLSACETMGSASCICTDKTGTLTT 1546
Query: 105 GRPVVTKV-VASTCIE--NANSSQTIENALSDVEILRLAAAVESNSVHPVGK------AI 155
VV K+ + +E S+ +++ +SD + L A+ N+ V K I
Sbjct: 1547 NHMVVDKIWICEKTVEMKGDESTDKLKSEISDEVLSILLQAIFQNTSSEVVKDNEGKQTI 1726
Query: 156 VDAAQAVNCLDAKVVDGTFLEEPGSGA----VATIGNRKVYVGTLEWITRHGINNNILQE 211
+ L+ +V G + V + + + L + G+
Sbjct: 1727 LGTPTESALLEFGLVSGGDFDAQRRSCKVLKVEPFNSDRKKMSVLVGLPDGGV------R 1888
Query: 212 VECKNESFVYVGVNDTL----AGLIYFEDEVREDARHVVDTLSKQDISVYML-------S 260
CK S + + + D + I +E ++D + + + L +
Sbjct: 1889 AFCKGASEIVLKMCDKIIDSNGTTIDLPEEKARIVSDIIDGFANEALRTLCLAVKDIDET 2068
Query: 261 GDKRNAAEH---VASLVGIPKDKVLSGVKPDQKKKFINELQKDNIVAMVGDGINDAAALA 317
+ N E+ + ++VGI KD V GVK +K + ++ + GD IN A A+
Sbjct: 2069 QGETNIPENGYTLITIVGI-KDPVRPGVKEAVQKCLAAGI---SVRMVTGDNINTAKAI- 2233
Query: 318 ASHIGIALGGGV---GAASEVSSIILMRD---HLSQLLDALELSRLTMTTVKQNLW 367
A GI GGV G S M+D + + +L L + T+ T +N++
Sbjct: 2234 AKECGILTEGGVAIEGPEFRNLSEEQMKDIIPRIQVMARSLPLDKHTLVTRLRNMF 2401
>TC85483 type IIB calcium ATPase MCA5 [Medicago truncatula]
Length = 2541
Score = 61.6 bits (148), Expect = 6e-10
Identities = 45/179 (25%), Positives = 80/179 (44%), Gaps = 28/179 (15%)
Frame = +2
Query: 227 TLAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVK 286
T G++ +D VR + V I+V M++GD N A+ +A GI D ++
Sbjct: 1004 TCIGVVGIKDPVRPGVKESVALCRSAGITVRMVTGDNINTAKAIARECGILTDDGIAIEG 1183
Query: 287 PDQKKKFINELQK---------------------------DNIVAMVGDGINDAAALAAS 319
P+ ++K + EL + +VA+ GDG NDA AL +
Sbjct: 1184 PEFREKSLEELLELIPKIQVMARSSPLDKHTLVRHLRTTFGEVVAVTGDGTNDAPALHEA 1363
Query: 320 HIGIALG-GGVGAASEVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGI 377
IG+A+G G A E + +I++ D+ S ++ + R +++ + + NIV +
Sbjct: 1364 DIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNIVAL 1540
Score = 36.6 bits (83), Expect = 0.020
Identities = 35/134 (26%), Positives = 53/134 (39%), Gaps = 3/134 (2%)
Frame = +2
Query: 8 GVMAVSVTTFTFWSLFGTHILPATAYQGSAVSLALQFACSVLVVACPCALGLATPTAVLV 67
G++++ + FW+ G L Y AV+ ++VVA P L LA ++
Sbjct: 185 GLVSLKLQQENFWNWNGDDALEMLEYFAIAVT--------IVVVAVPEGLPLAVTLSLAF 340
Query: 68 GTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKV---VASTCIENANSS 124
L+R E + DKTGTLT V K + S + N SS
Sbjct: 341 AMKKMMNDKALVRNLAACETMGSATTICSDKTGTLTTNHMTVVKTCICMKSKEVSNKTSS 520
Query: 125 QTIENALSDVEILR 138
E S V++L+
Sbjct: 521 LCSELPESVVKLLQ 562
>TC92182 type IIA calcium ATPase [Medicago truncatula]
Length = 3148
Score = 61.2 bits (147), Expect = 8e-10
Identities = 46/178 (25%), Positives = 78/178 (42%), Gaps = 33/178 (18%)
Frame = +1
Query: 230 GLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVAS---LVGIPKD------- 279
G++ D RE+ ++ + I V +++GD ++ AE + L +D
Sbjct: 1843 GVVGLRDPPREEVHKAIEDCKQAGIRVMVITGDNKSTAEAICKEIKLFSTDEDLTGQSLT 2022
Query: 280 ---------------------KVLSGVKPDQKKKFINELQK-DNIVAMVGDGINDAAALA 317
KV S +P K++ + L++ IVAM GDG+NDA AL
Sbjct: 2023 GKEFMSLSHSEQVKLLLRNGGKVFSRAEPRHKQEIVRLLKEMGEIVAMTGDGVNDAPALK 2202
Query: 318 ASHIGIALG-GGVGAASEVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNI 374
+ IGIA+G G A E S ++L D+ S ++ A+ R +K + + N+
Sbjct: 2203 LADIGIAMGITGTEVAKEASDMVLADDNFSTIVSAIAEGRAIYNNMKAFIRYMISSNV 2376
Score = 32.3 bits (72), Expect = 0.38
Identities = 19/69 (27%), Positives = 32/69 (45%)
Frame = +1
Query: 43 QFACSVLVVACPCALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTL 102
+ A ++ V A P L T + +GT A++ ++R +E + DKTGTL
Sbjct: 925 KIAVALAVAAIPEGLPAVITTCLALGTRKMAQKNAIVRKLPSVETLGCTTVICSDKTGTL 1104
Query: 103 TVGRPVVTK 111
T + T+
Sbjct: 1105TTNQMSATE 1131
>BQ138304 homologue to GP|6688833|emb putative calcium P-type ATPase
{Neurospora crassa}, partial (13%)
Length = 412
Score = 58.9 bits (141), Expect = 4e-09
Identities = 32/95 (33%), Positives = 52/95 (54%), Gaps = 1/95 (1%)
Frame = +3
Query: 281 VLSGVKPDQKKKFINELQK-DNIVAMVGDGINDAAALAASHIGIALGGGVGAASEVSSII 339
+ S +P K K ++ LQK +VAM GDG+NDA AL + IG+A+G G A + ++
Sbjct: 39 LFSRTEPTHKSKLVDLLQKAGEVVAMTGDGVNDAPALKKADIGVAMGSGTDVAKLAADMV 218
Query: 340 LMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNI 374
L+ D+ + + A+E R +Q + + NI
Sbjct: 219 LVDDNFATIEGAVEEGRSIYNNTQQFIRYLISSNI 323
>BF640720 homologue to PIR|S52728|S52 H+-transporting ATPase (EC 3.6.1.35) -
kidney bean, partial (24%)
Length = 689
Score = 57.8 bits (138), Expect = 9e-09
Identities = 36/104 (34%), Positives = 52/104 (49%), Gaps = 1/104 (0%)
Frame = +2
Query: 283 SGVKPDQKKKFINELQ-KDNIVAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILM 341
+GV P+ K + + LQ K +I M GDG+NDA AL + IGIA+ AA S I+L
Sbjct: 8 AGVFPEHKYEIVKRLQEKKHICGMTGDGVNDAPALKKADIGIAVADATDAARSASDIVLT 187
Query: 342 RDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGIPIAAGVLF 385
LS ++ A+ SR +K Y I + I ++F
Sbjct: 188 EPGLSVIISAVLTSRAIFQRMKN-------YTIYAVSITIRIVF 298
>TC87649 similar to SP|Q9SZR1|ACAA_ARATH Potential calcium-transporting
ATPase 10 plasma membrane-type (EC 3.6.3.8), partial
(43%)
Length = 2159
Score = 56.2 bits (134), Expect = 2e-08
Identities = 43/184 (23%), Positives = 79/184 (42%), Gaps = 32/184 (17%)
Frame = +3
Query: 228 LAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVA-------SLVGIPKDK 280
L ++ +D R ++ V K + V M++GD A+ +A SL + +
Sbjct: 351 LLAIVGIKDPCRPGVKNSVQLCQKAGVKVKMVTGDNVKTAKAIALECGILSSLADVTERS 530
Query: 281 VLSGV-----------------------KPDQKKKFINELQ-KDNIVAMVGDGINDAAAL 316
V+ G P+ K + L+ K ++VA+ GDG NDA AL
Sbjct: 531 VIEGKTFRALSDSEREEIAESISVMGRSSPNDKLLLVQALRRKGHVVAVTGDGTNDAPAL 710
Query: 317 AASHIGIALG-GGVGAASEVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIV 375
+ IG+A+G G A E S II++ D+ + ++ + R +++ + + N+
Sbjct: 711 HEADIGLAMGIAGTEVAKESSDIIILDDNFASVVKVVRWGRSVYANIQKFIQFQLTVNVA 890
Query: 376 GIPI 379
+ I
Sbjct: 891 ALVI 902
>BF634181 homologue to SP|Q08436|PMA3 Plasma membrane ATPase 3 (EC 3.6.3.6)
(Proton pump 3). [Leadwort-leaved tobacco], partial
(23%)
Length = 663
Score = 55.1 bits (131), Expect = 6e-08
Identities = 33/89 (37%), Positives = 47/89 (52%), Gaps = 1/89 (1%)
Frame = +3
Query: 276 IPKDKVLSGVKPDQKKKFINELQ-KDNIVAMVGDGINDAAALAASHIGIALGGGVGAASE 334
I K +GV P+ K + + LQ + +I M GDG+NDA AL + IGIA+ AA
Sbjct: 159 IEKADGFAGVFPEHKYEIVKRLQARKHICGMTGDGVNDAPALKKADIGIAVDDATDAARS 338
Query: 335 VSSIILMRDHLSQLLDALELSRLTMTTVK 363
S I+L LS ++ A+ SR +K
Sbjct: 339 ASDIVLTEPGLSVIISAVLTSRAIFQRMK 425
>TC88464 similar to SP|Q9LU41|ACA9_ARATH Potential calcium-transporting
ATPase 9 plasma membrane-type (EC 3.6.3.8) (Ca2+-ATPase
isoform 9)., partial (43%)
Length = 1608
Score = 51.6 bits (122), Expect = 6e-07
Identities = 41/184 (22%), Positives = 76/184 (41%), Gaps = 32/184 (17%)
Frame = +3
Query: 228 LAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGI----------- 276
L ++ +D R + V + + V M++GD A+ +A GI
Sbjct: 258 LLAIVGIKDPCRPGVKDAVRICTDAGVKVRMVTGDNLQTAKAIALECGILASNEEAVDPC 437
Query: 277 -------------------PKDKVLSGVKPDQKKKFINELQKD-NIVAMVGDGINDAAAL 316
K V+ P+ K + L+K ++VA+ GDG NDA AL
Sbjct: 438 IIEGKVFRELSEKEREQVAKKITVMGRSSPNDKLLLVQALRKGGDVVAVTGDGTNDAPAL 617
Query: 317 AASHIGIALG-GGVGAASEVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIV 375
+ IG+++G G A E S II++ D+ + ++ + R +++ + + N+
Sbjct: 618 HEADIGLSMGIQGTEVAKESSDIIILDDNFASVVKVVRWGRSVYANIQKFIQFQLTVNVA 797
Query: 376 GIPI 379
+ I
Sbjct: 798 ALVI 809
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,485,314
Number of Sequences: 36976
Number of extensions: 140562
Number of successful extensions: 756
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 732
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 743
length of query: 451
length of database: 9,014,727
effective HSP length: 99
effective length of query: 352
effective length of database: 5,354,103
effective search space: 1884644256
effective search space used: 1884644256
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0182.6


