

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
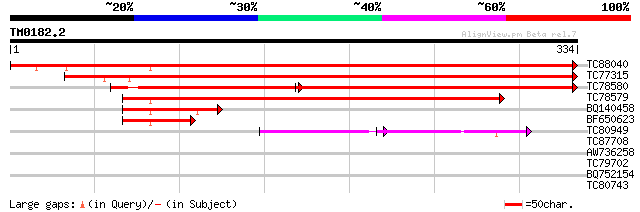
Query= TM0182.2
(334 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88040 homologue to SP|Q41266|AOX2_SOYBN Alternative oxidase 2 ... 549 e-157
TC77315 homologue to PIR|A46364|A46364 alternative respiratory p... 432 e-122
TC78580 similar to SP|O03376|AOX3_SOYBN Alternative oxidase 3 m... 295 e-119
TC78579 similar to GP|17154769|emb|CAD12835. putative alternativ... 367 e-102
BQ140458 similar to GP|17154769|emb putative alternative oxidase... 89 2e-18
BF650623 similar to GP|17154769|emb putative alternative oxidase... 70 1e-12
TC80949 similar to PIR|T52422|T52422 alternative oxidase-related... 51 1e-11
TC87708 homologue to PIR|T06680|T06680 hypothetical protein T17F... 30 1.7
AW736258 similar to PIR|T46090|T46 hypothetical protein T20E23.1... 28 3.8
TC79702 MtN14 28 4.9
BQ752154 27 8.4
TC80743 similar to GP|20856938|gb|AAM26691.1 At1g73960/F2P9_17 {... 27 8.4
>TC88040 homologue to SP|Q41266|AOX2_SOYBN Alternative oxidase 2
mitochondrial precursor (EC 1.-.-.-). [Soybean] {Glycine
max}, partial (82%)
Length = 1678
Score = 549 bits (1415), Expect = e-157
Identities = 273/345 (79%), Positives = 289/345 (83%), Gaps = 11/345 (3%)
Frame = +2
Query: 1 MKHLALSYALRRAL----NCNRHGLTAVRQLPATEV--RRFLVSGENGVFSCWNRMMSSQ 54
MKH AL Y RRAL N NR VR A E+ + G NG W RMMSSQ
Sbjct: 392 MKHSALCYVARRALIGGRNSNRQSSAVVRSFAAAEIGQKHLYADGGNGGLFYWKRMMSSQ 571
Query: 55 AAPEEEKKEEKAEKESLRTEAKKNDGS-----VVVSSYWGISRPKITREDGTEWPWNCFM 109
AAP + EE K + + E KK + S VV SSYWGISRPKI REDGTEWPWNCFM
Sbjct: 572 AAPSKPSAEETEAKSTEKNEKKKEESSGTKNNVVASSYWGISRPKIMREDGTEWPWNCFM 751
Query: 110 PWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVP 169
PWETY+ ++SIDL KHHVPKNFLDKVAYRTVKLLRIPTDVFF+RRYGCRAMMLETVAAVP
Sbjct: 752 PWETYQSNVSIDLNKHHVPKNFLDKVAYRTVKLLRIPTDVFFKRRYGCRAMMLETVAAVP 931
Query: 170 GMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGV 229
GMVGGMLLHL+SLRKFQ SGGW+KALLEEAENERMHLMTMVELV+PKWYERFLVL VQGV
Sbjct: 932 GMVGGMLLHLKSLRKFQHSGGWVKALLEEAENERMHLMTMVELVKPKWYERFLVLAVQGV 1111
Query: 230 FFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPK 289
FFNAFFVLY+LSPKVAHRVVGYLEEEAIHSYTEYLKDI+SGAIENVPAPAIAIDYWRLPK
Sbjct: 1112FFNAFFVLYILSPKVAHRVVGYLEEEAIHSYTEYLKDIDSGAIENVPAPAIAIDYWRLPK 1291
Query: 290 DATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
DA LKDVITVIRADEAHHRDVNHFASDIHFHGKELR+APAPLGYH
Sbjct: 1292DAKLKDVITVIRADEAHHRDVNHFASDIHFHGKELRDAPAPLGYH 1426
>TC77315 homologue to PIR|A46364|A46364 alternative respiratory pathway
oxidase (EC 1.-.-.-) - Arabidopsis thaliana, partial
(84%)
Length = 1495
Score = 432 bits (1111), Expect = e-122
Identities = 207/314 (65%), Positives = 251/314 (79%), Gaps = 12/314 (3%)
Frame = +1
Query: 33 RRFLVSGENGVFSCWNRMMSSQ----------AAPEEEKKEEKAEKE--SLRTEAKKNDG 80
++ L+ GE GV + W ++ S A ++K + +K S + A N
Sbjct: 142 KKGLLGGEVGVPNKWGYLVRSTPLVRKTSTFTANLSDQKDNKNVDKTPPSSQGGAGDNKD 321
Query: 81 SVVVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLSIDLTKHHVPKNFLDKVAYRTV 140
++SYWG+ KIT+ DGTEW WNCF PWETY+ D++IDLTKHH P FLDK+AY TV
Sbjct: 322 EKGITSYWGVQPSKITKPDGTEWKWNCFRPWETYKADVTIDLTKHHKPTTFLDKMAYWTV 501
Query: 141 KLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAE 200
K LR PTD+FFQRRYGCRAMMLETVAAVPGMVGGMLLH +SLR+F+QSGGWIKALLEEAE
Sbjct: 502 KSLRWPTDIFFQRRYGCRAMMLETVAAVPGMVGGMLLHCKSLRRFEQSGGWIKALLEEAE 681
Query: 201 NERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSY 260
NERMHLMT +E+ +PKWYER LV+TVQGVFFNA+F+ YLLSPK AHR+VGYLEEEAIHSY
Sbjct: 682 NERMHLMTFMEVAKPKWYERALVITVQGVFFNAYFLGYLLSPKFAHRMVGYLEEEAIHSY 861
Query: 261 TEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIHFH 320
TE+LK+++ G IENVPAPAIAIDYW+LP+++TL+DV+ V+RADEAHHRDVNHFASDIH+
Sbjct: 862 TEFLKELDKGNIENVPAPAIAIDYWQLPQNSTLRDVVEVVRADEAHHRDVNHFASDIHYQ 1041
Query: 321 GKELREAPAPLGYH 334
G+ELREA AP+GYH
Sbjct: 1042GRELREAAAPIGYH 1083
>TC78580 similar to SP|O03376|AOX3_SOYBN Alternative oxidase 3
mitochondrial precursor (EC 1.-.-.-). [Soybean] {Glycine
max}, partial (87%)
Length = 992
Score = 295 bits (755), Expect(2) = e-119
Identities = 140/166 (84%), Positives = 152/166 (91%)
Frame = +3
Query: 169 PGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQG 228
P MVGGMLLHL+SLRKFQ +GGWIKALLEEAENERMHLMTMVELV+P W+ER LV+T QG
Sbjct: 372 PPMVGGMLLHLKSLRKFQHTGGWIKALLEEAENERMHLMTMVELVKPSWHERLLVITAQG 551
Query: 229 VFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLP 288
VFFNAFFV Y+LSPK AHR VGYLEEEA+ SYT++L IESG +ENVPAPAIAIDYWRLP
Sbjct: 552 VFFNAFFVFYILSPKTAHRFVGYLEEEAVISYTQHLNAIESGKVENVPAPAIAIDYWRLP 731
Query: 289 KDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
KDATLKDVITVIRADEAHHRDVNHFASDIH GKEL+EAPAP+GYH
Sbjct: 732 KDATLKDVITVIRADEAHHRDVNHFASDIHHQGKELKEAPAPIGYH 869
Score = 152 bits (383), Expect(2) = e-119
Identities = 69/114 (60%), Positives = 86/114 (74%)
Frame = +2
Query: 60 EKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLS 119
EKK++ +E+ ND + VVSSYWGI+RPK+ REDGTEWPWNC MPWE+Y D+S
Sbjct: 59 EKKDQHSEENK-----NSNDSNTVVSSYWGITRPKVKREDGTEWPWNCGMPWESYSSDVS 223
Query: 120 IDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVG 173
ID+TKHHVPK F DK A+R+VK LR+ +D+ F+ RYGC AMMLET+AA P G
Sbjct: 224 IDVTKHHVPKTFGDKFAFRSVKFLRVLSDLDFKERYGCHAMMLETIAARPANGG 385
>TC78579 similar to GP|17154769|emb|CAD12835. putative alternative oxidase
{Vigna unguiculata}, partial (66%)
Length = 975
Score = 367 bits (942), Expect = e-102
Identities = 170/227 (74%), Positives = 196/227 (85%), Gaps = 2/227 (0%)
Frame = +2
Query: 67 EKESLRTEAKKNDGS--VVVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLSIDLTK 124
EK+ +TE K D + VVSSYWGISRPK+ +EDGTEWPWNCFMPWE+Y D+SID+TK
Sbjct: 293 EKKDQQTEESKKDANHNAVVSSYWGISRPKVLKEDGTEWPWNCFMPWESYSSDVSIDVTK 472
Query: 125 HHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRK 184
HHVPK F DK A+R+VK LR+ +D++F+ RYGC AMMLET+AAVPGMVGGMLLHL+SLRK
Sbjct: 473 HHVPKTFGDKFAFRSVKFLRVLSDLYFKERYGCHAMMLETIAAVPGMVGGMLLHLKSLRK 652
Query: 185 FQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYLLSPKV 244
FQ +GGWIKALLEEAENERMHLMTMVELV+P W+ER LV+T QGVFFN FFV Y+LSPK+
Sbjct: 653 FQHAGGWIKALLEEAENERMHLMTMVELVKPSWHERLLVITAQGVFFNGFFVFYILSPKI 832
Query: 245 AHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDA 291
AHR VGYLEEEA+ SYT+YL IESG +ENVPAPAIAIDYWRLP DA
Sbjct: 833 AHRFVGYLEEEAVISYTQYLNAIESGKVENVPAPAIAIDYWRLPNDA 973
>BQ140458 similar to GP|17154769|emb putative alternative oxidase {Vigna
unguiculata}, partial (24%)
Length = 384
Score = 89.0 bits (219), Expect = 2e-18
Identities = 41/69 (59%), Positives = 49/69 (70%), Gaps = 10/69 (14%)
Frame = +3
Query: 67 EKESLRTEAKKNDGS--VVVSSYWGISRPKITREDGTEWPWNCFM--------PWETYRP 116
EK+ +TE K D + VVSSYWGISRPK+ +EDGTEWPWNCFM PWE+Y
Sbjct: 177 EKKDQQTEESKKDANHNAVVSSYWGISRPKVLKEDGTEWPWNCFMVRIHI*KEPWESYXS 356
Query: 117 DLSIDLTKH 125
D+SID+TKH
Sbjct: 357 DVSIDVTKH 383
>BF650623 similar to GP|17154769|emb putative alternative oxidase {Vigna
unguiculata}, partial (19%)
Length = 647
Score = 69.7 bits (169), Expect = 1e-12
Identities = 30/45 (66%), Positives = 35/45 (77%), Gaps = 2/45 (4%)
Frame = +3
Query: 67 EKESLRTEAKKNDGS--VVVSSYWGISRPKITREDGTEWPWNCFM 109
EK+ +TE K D + VVSSYWGISRPK+ +EDGTEWPWNCFM
Sbjct: 183 EKKDQQTEESKKDANHNAVVSSYWGISRPKVLKEDGTEWPWNCFM 317
>TC80949 similar to PIR|T52422|T52422 alternative oxidase-related protein
IMMUTANS [validated] - Arabidopsis thaliana, partial
(69%)
Length = 1369
Score = 51.2 bits (121), Expect(2) = 1e-11
Identities = 29/110 (26%), Positives = 54/110 (48%), Gaps = 19/110 (17%)
Frame = +3
Query: 217 WYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVP 276
W++RFL + ++ ++YL+SP++A+ +E A +Y +++K+ + ++ +P
Sbjct: 591 WFDRFLAQHIAIFYYFMTALMYLISPRMAYHFSECVESHAFETYDKFIKE-QGEELKKMP 767
Query: 277 APAIAIDYW-------------------RLPKDATLKDVITVIRADEAHH 307
AP +A++Y+ R P L DV IR DEA H
Sbjct: 768 APEVAVNYYTGGDLYLFDEFQTSRVPNTRRPTIDNLYDVFLNIRDDEAEH 917
Score = 35.4 bits (80), Expect(2) = 1e-11
Identities = 23/76 (30%), Positives = 36/76 (47%)
Frame = +2
Query: 148 DVFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLM 207
D ++ R R +LET+A VP +LH+ + + ++K E+ NE HL+
Sbjct: 380 DALYRDRNYARFFVLETIARVPYFAFMSILHMYESFGWWRRADYLKVHFAESWNEMHHLL 559
Query: 208 TMVELVQPKWYERFLV 223
M EL W E +V
Sbjct: 560 IMEEL----WRECLVV 595
>TC87708 homologue to PIR|T06680|T06680 hypothetical protein T17F15.100 -
Arabidopsis thaliana, partial (16%)
Length = 805
Score = 29.6 bits (65), Expect = 1.7
Identities = 17/43 (39%), Positives = 24/43 (55%)
Frame = +2
Query: 111 WETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQR 153
W +Y +L + L HH P F+ R+ +LL IPT +FF R
Sbjct: 122 WPSYPHNLHLFLLFHH-PIKFI----VRSDRLLNIPTTIFFDR 235
>AW736258 similar to PIR|T46090|T46 hypothetical protein T20E23.190 -
Arabidopsis thaliana, partial (21%)
Length = 618
Score = 28.5 bits (62), Expect = 3.8
Identities = 15/37 (40%), Positives = 18/37 (48%)
Frame = +1
Query: 35 FLVSGENGVFSCWNRMMSSQAAPEEEKKEEKAEKESL 71
F VS EN +SCWN S A + + EE K L
Sbjct: 259 FWVS*ENECYSCWNG*RRSNAYQNQTRPEEGYRKSCL 369
>TC79702 MtN14
Length = 973
Score = 28.1 bits (61), Expect = 4.9
Identities = 20/58 (34%), Positives = 26/58 (44%), Gaps = 1/58 (1%)
Frame = +2
Query: 23 AVRQLPATEVRRF-LVSGENGVFSCWNRMMSSQAAPEEEKKEEKAEKESLRTEAKKND 79
AV++ PA E +V EN V + EEK EEK EKE E K+ +
Sbjct: 401 AVQEKPAEESETVNVVKDENVVAEPETKDNVKTEETSEEKNEEKVEKEDAMDEKKEEE 574
>BQ752154
Length = 674
Score = 27.3 bits (59), Expect = 8.4
Identities = 11/31 (35%), Positives = 14/31 (44%)
Frame = -1
Query: 103 WPWNCFMPWETYRPDLSIDLTKHHVPKNFLD 133
WPW F PW + T HH+P +D
Sbjct: 437 WPWTTFPPW--------MVRTGHHIPNLAID 369
>TC80743 similar to GP|20856938|gb|AAM26691.1 At1g73960/F2P9_17 {Arabidopsis
thaliana}, partial (17%)
Length = 1236
Score = 27.3 bits (59), Expect = 8.4
Identities = 13/33 (39%), Positives = 18/33 (54%)
Frame = -2
Query: 35 FLVSGENGVFSCWNRMMSSQAAPEEEKKEEKAE 67
FLVSG N +S WN ++ A P+E + E
Sbjct: 281 FLVSGSNRSYSVWNGLL---ALPQERNTRQNRE 192
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.136 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,730,093
Number of Sequences: 36976
Number of extensions: 140881
Number of successful extensions: 836
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 811
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 826
length of query: 334
length of database: 9,014,727
effective HSP length: 97
effective length of query: 237
effective length of database: 5,428,055
effective search space: 1286449035
effective search space used: 1286449035
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0182.2


