

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
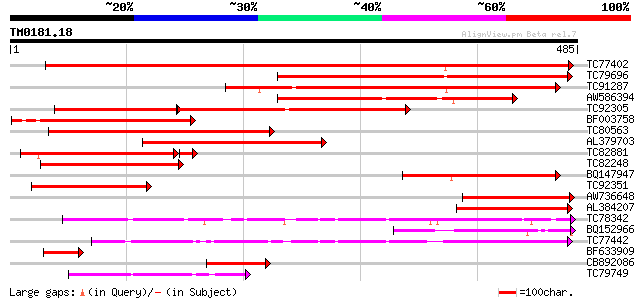
Query= TM0181.18
(485 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77402 similar to PIR|T50691|T50691 amino acid permease 6 [impo... 583 e-167
TC79696 similar to GP|15216028|emb|CAC51424. amino acid permease... 352 2e-97
TC91287 weakly similar to GP|10176848|dbj|BAB10054. amino acid t... 316 1e-86
AW586394 similar to GP|15216026|em amino acid permease AAP1 {Vic... 307 7e-84
TC92305 weakly similar to GP|10176848|dbj|BAB10054. amino acid t... 207 9e-81
BF003758 similar to GP|15216030|em amino acid permease AAP4 {Vic... 263 1e-70
TC80563 weakly similar to GP|10176848|dbj|BAB10054. amino acid t... 189 3e-48
AL379703 homologue to GP|6539600|gb| amino acid transporter a {V... 188 3e-48
TC82881 similar to GP|15216026|emb|CAC51423. amino acid permease... 181 7e-47
TC82248 similar to GP|15216028|emb|CAC51424. amino acid permease... 174 9e-44
BQ147947 weakly similar to GP|10176848|db amino acid transporter... 148 4e-36
TC92351 homologue to GP|6539604|gb|AAF15946.1| amino acid transp... 122 2e-28
AW736648 similar to PIR|T50691|T506 amino acid permease 6 [impor... 120 2e-27
AL384207 weakly similar to GP|21553710|gb| amino acid carrier p... 115 5e-26
TC78342 similar to GP|8571474|gb|AAF76897.1| proline/glycine bet... 95 5e-20
BQ152966 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsi... 73 2e-13
TC77442 similar to GP|20804928|dbj|BAB92607. proline transport p... 71 1e-12
BF633909 similar to GP|13676299|gb| amino acid transporter {Glyc... 69 3e-12
CB892086 similar to GP|6539604|gb|A amino acid transporter c {Vi... 66 3e-11
TC79749 weakly similar to GP|15290010|dbj|BAB63704. contains EST... 58 7e-09
>TC77402 similar to PIR|T50691|T50691 amino acid permease 6 [imported] -
Arabidopsis thaliana, partial (90%)
Length = 1902
Score = 583 bits (1504), Expect = e-167
Identities = 275/456 (60%), Positives = 355/456 (77%), Gaps = 4/456 (0%)
Frame = +3
Query: 31 KCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYT 90
K FDDDGR KRTGT TASAHIITAVIGSGVLSLAWAIAQ+GWVAGPAV+ FS +TY+T
Sbjct: 330 KNFDDDGRAKRTGTWLTASAHIITAVIGSGVLSLAWAIAQMGWVAGPAVLFAFSFITYFT 509
Query: 91 SILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSIS 150
S LL CYR+ DPV+GKRNYTY +VV +N+GG + +LCG+ QY+NL GV IGYTI +SIS
Sbjct: 510 STLLADCYRSPDPVHGKRNYTYTEVVRANLGGRKFQLCGLAQYINLVGVTIGYTITASIS 689
Query: 151 MIAIERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTY 210
M+A++RSNCFHK +D C+++ N +MI F ++IVL QIP+F +L WLSI+AAVMSF Y
Sbjct: 690 MVAVQRSNCFHKHGHQDKCYVSNNPFMIIFACIQIVLCQIPNFHELSWLSIVAAVMSFAY 869
Query: 211 STIGLGLGFGKVIENG-AVGGSLTGITVGT-VTQTQKVWRTLQALGDIAFAYSYSMILIE 268
S+IGLGL KV G V SLTG+ +G VT T+KVWR QA+GDIAFAY++S +LIE
Sbjct: 870 SSIGLGLSVAKVAGGGNHVTTSLTGVQIGVDVTATEKVWRMFQAIGDIAFAYAFSNVLIE 1049
Query: 269 IQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFW 328
IQDT+KS P E++ MK+AS I ++ TTLFY+LCG GYAAFG+ +PGN LTGFGFY PFW
Sbjct: 1050IQDTLKSSPPENRVMKRASLIGILTTTLFYVLCGTLGYAAFGNDAPGNFLTGFGFYEPFW 1229
Query: 329 LIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVT--KTVQIPLFNAYKINL 386
LID AN I +HL+GAYQV+ QP+F FVE + +K+PDS FV + IPL+ +Y +N
Sbjct: 1230LIDFANVCIAVHLIGAYQVFVQPIFGFVEGQSKQKWPDSKFVNGEHAMNIPLYGSYNVNY 1409
Query: 387 FRLVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKW 446
FR++WR+ +VIIT +I+ML PFFND +GL+G+L F+PLTVYFP+EMYI + +PK+S W
Sbjct: 1410FRVIWRSCYVIITAIIAMLFPFFNDFLGLIGSLSFYPLTVYFPIEMYIKKTNMPKYSFTW 1589
Query: 447 ICFQLFSLACLIVTVAAAAGSVAGIFLDLQVYKPFK 482
++ S CL++++ +AAGS+ G+ L+ YKPF+
Sbjct: 1590TWLKILSWLCLVISIISAAGSIQGLATSLKTYKPFR 1697
>TC79696 similar to GP|15216028|emb|CAC51424. amino acid permease AAP3
{Vicia faba var. minor}, partial (52%)
Length = 1111
Score = 352 bits (903), Expect = 2e-97
Identities = 166/252 (65%), Positives = 203/252 (79%)
Frame = +2
Query: 230 GSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESKTMKKASFI 289
GSLTG++VGTVT QKVW Q LG+IAFAYSYS +L+EIQDT+KSPPSE K MK A+ I
Sbjct: 5 GSLTGVSVGTVTPAQKVWGVFQGLGNIAFAYSYSFVLLEIQDTIKSPPSEGKAMKIAAKI 184
Query: 290 SVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYS 349
S+ VTT FY+LCGC GYAAFGD++PGNLL GFG +W++D ANAAIVIHL GAYQVY+
Sbjct: 185 SIAVTTTFYLLCGCMGYAAFGDNAPGNLLAGFGVSKAYWVVDAANAAIVIHLFGAYQVYA 364
Query: 350 QPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWRTIFVIITTLISMLLPFF 409
QPLFAFVE AA+K+P D K V+IP Y N+F LVWRT+FVII+TLI+ML+PFF
Sbjct: 365 QPLFAFVEKEAAKKWPKIDREFK-VKIPGLPVYSQNIFSLVWRTVFVIISTLIAMLIPFF 541
Query: 410 NDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLACLIVTVAAAAGSVA 469
ND++G++GAL FWPLTVYFPVEMYI+Q +IPKWS KWI ++ S CLIV++ A GS+
Sbjct: 542 NDVLGVIGALXFWPLTVYFPVEMYIVQMKIPKWSRKWIILEIMSTFCLIVSIVAGLGSLV 721
Query: 470 GIFLDLQVYKPF 481
G+++DLQ YKPF
Sbjct: 722 GVWIDLQKYKPF 757
>TC91287 weakly similar to GP|10176848|dbj|BAB10054. amino acid transporter
{Arabidopsis thaliana}, partial (43%)
Length = 919
Score = 316 bits (810), Expect = 1e-86
Identities = 150/291 (51%), Positives = 209/291 (71%), Gaps = 4/291 (1%)
Frame = +1
Query: 185 IVLSQIPDFDQLWWLSILAAVMSFTYST--IGLGLGFGKVIENGAVGGSLTGITVGTVTQ 242
IV++ IPD + L+A T T IGL LG VI+NG + GS+TG+ V
Sbjct: 4 IVMAIIPDLHNMALGFHLSAST*CTLHTSFIGL*LGLSTVIKNGRIMGSITGLQKAKVAD 183
Query: 243 TQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCG 302
K+W QA+GDI +Y YS+IL+EIQDT++SPP E++TMKKAS +++ +TT FY+ CG
Sbjct: 184 --KIWLIFQAIGDINLSYPYSIILLEIQDTLESPPPENQTMKKASMVAIFITTFFYLCCG 357
Query: 303 CFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAE 362
CFGYAAFGD++PGNLLTGFGF+ P+WLIDIAN I+IHLVG YQ+YSQP+++ + + +
Sbjct: 358 CFGYAAFGDATPGNLLTGFGFFEPYWLIDIANVCIIIHLVGGYQIYSQPIYSTADRWFTK 537
Query: 363 KFPDSDFVTK--TVQIPLFNAYKINLFRLVWRTIFVIITTLISMLLPFFNDIVGLLGALG 420
K+P+S FV V++PL +++INLFR +RT +VI TT +++L P+FN ++GLLGA+
Sbjct: 538 KYPNSGFVNNFHKVKLPLLPSFEINLFRFCFRTSYVISTTGLAILFPYFNSVLGLLGAIN 717
Query: 421 FWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLACLIVTVAAAAGSVAGI 471
FWPL +YFPVEMY +QK++ W+ KWI ++FS AC +VT+ GS GI
Sbjct: 718 FWPLAIYFPVEMYFVQKKVGAWTRKWIVLRIFSFACFLVTMVGFVGSFEGI 870
>AW586394 similar to GP|15216026|em amino acid permease AAP1 {Vicia faba var.
minor}, partial (42%)
Length = 613
Score = 307 bits (786), Expect = 7e-84
Identities = 148/207 (71%), Positives = 178/207 (85%), Gaps = 2/207 (0%)
Frame = +1
Query: 230 GSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESKTMKKASFI 289
GSLTG+++GTVT+ QKVW T QALG+IAFAYSYS ILIEIQDT+K+PPSE KTMK+A+ I
Sbjct: 4 GSLTGVSIGTVTKAQKVWGTFQALGNIAFAYSYSQILIEIQDTIKNPPSEVKTMKQATKI 183
Query: 290 SVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYS 349
S+ VTT FYMLCGC GYAAFGD++PGNLLT G +NP+WLIDIANAAIVIHLVGAYQVY+
Sbjct: 184 SIGVTTAFYMLCGCMGYAAFGDTAPGNLLT--GIFNPYWLIDIANAAIVIHLVGAYQVYA 357
Query: 350 QPLFAFVENYAAEKFPDSDFVTKTVQIPL--FNAYKINLFRLVWRTIFVIITTLISMLLP 407
QP FAFVE +++P + K +IP+ F+ Y +NLFRL+WRTIFVI TT+I+ML+P
Sbjct: 358 QPFFAFVEKIVIKRWPK---INKEYRIPIPGFHPYNLNLFRLIWRTIFVITTTVIAMLIP 528
Query: 408 FFNDIVGLLGALGFWPLTVYFPVEMYI 434
FFND++GLLGA+GFWPLTVYFPVEMYI
Sbjct: 529 FFNDVLGLLGAVGFWPLTVYFPVEMYI 609
>TC92305 weakly similar to GP|10176848|dbj|BAB10054. amino acid transporter
{Arabidopsis thaliana}, partial (43%)
Length = 1048
Score = 207 bits (527), Expect(2) = 9e-81
Identities = 102/200 (51%), Positives = 142/200 (71%)
Frame = +2
Query: 144 TIASSISMIAIERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILA 203
++ +IS+ AI+ S H E + P YM+ FG+V+I LSQIP+ + WLS++A
Sbjct: 455 SLQXAISLRAIQISISQHNKENETPSEFADAYYMLIFGIVQIALSQIPNLHDIHWLSVVA 634
Query: 204 AVMSFTYSTIGLGLGFGKVIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYS 263
A+ SF Y IG+GL ++IENG GS+ GI+ T + T+K+W QALGD++F+Y +S
Sbjct: 635 AITSFGYWFIGMGLSIMQIIENGYAKGSIEGIS--TSSGTEKLWLVSQALGDVSFSYPFS 808
Query: 264 MILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGF 323
I++EIQDT+K+PP E++TMKKAS ISV +TT FY++CG GYAAFGD++PGNLLTGFG
Sbjct: 809 TIMMEIQDTLKTPPPENQTMKKASTISVAITTFFYLVCGWAGYAAFGDNTPGNLLTGFGS 988
Query: 324 YNPFWLIDIANAAIVIHLVG 343
+WL+ A+A IV HLVG
Sbjct: 989 SKFYWLVGFAHACIVXHLVG 1048
Score = 111 bits (278), Expect(2) = 9e-81
Identities = 53/110 (48%), Positives = 77/110 (69%), Gaps = 1/110 (0%)
Frame = +1
Query: 39 LKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACY 98
+KRTGT+WTA AHI+T VIGSGVLSL W+IAQLGW+ GP ++L + T Y++ LLC Y
Sbjct: 136 IKRTGTLWTAVAHIVTGVIGSGVLSLPWSIAQLGWIVGPFSILLIASSTLYSAFLLCNTY 315
Query: 99 RNGDPVNG-KRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIAS 147
R+ +P G R+ +Y+DVV+ N+G +LCG + + ++G I + I +
Sbjct: 316 RSPNPEYGPHRSASYLDVVNFNLGTGNGRLCGFLVNICIYGFGIAFVITT 465
>BF003758 similar to GP|15216030|em amino acid permease AAP4 {Vicia faba var.
minor}, partial (31%)
Length = 554
Score = 263 bits (671), Expect = 1e-70
Identities = 130/158 (82%), Positives = 140/158 (88%)
Frame = +2
Query: 2 KMNEKNGSKNNHHHQAFDVSLDMQQQGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGV 61
KM EKNGS N HHQ FDVS+D QQ SK FDDDGR+KRTGT WTASAH+ITAVIGSGV
Sbjct: 92 KMVEKNGSTN--HHQTFDVSID--QQRDSKFFDDDGRVKRTGTAWTASAHVITAVIGSGV 259
Query: 62 LSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMG 121
LSLAWAIAQLGW+AGP VMILF+ VTYYTSILLC CYR GDP++GKRNYTYMDVVHSN+G
Sbjct: 260 LSLAWAIAQLGWIAGPIVMILFAWVTYYTSILLCECYRTGDPISGKRNYTYMDVVHSNLG 439
Query: 122 GIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNC 159
G QV LCGIVQYLNL GVAIGYTIAS+ISM+AIERSNC
Sbjct: 440 GFQVTLCGIVQYLNLVGVAIGYTIASAISMMAIERSNC 553
>TC80563 weakly similar to GP|10176848|dbj|BAB10054. amino acid transporter
{Arabidopsis thaliana}, partial (32%)
Length = 823
Score = 189 bits (479), Expect = 3e-48
Identities = 95/194 (48%), Positives = 132/194 (67%), Gaps = 1/194 (0%)
Frame = +2
Query: 34 DDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSIL 93
D+ +KRTGTVWTA AHI+T VIGSGVLSLAW+IAQ+GW+AGP +ILF+ VT ++ L
Sbjct: 242 DNSYHVKRTGTVWTAVAHIVTGVIGSGVLSLAWSIAQIGWIAGPLAIILFASVTLLSAFL 421
Query: 94 LCACYRNGDPVNG-KRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMI 152
L YR+ DP G +R+ +Y+D V+ + G +LCG+ ++L+G I Y I +SIS+
Sbjct: 422 LSDTYRSPDPELGPRRSSSYLDAVNIHKGEKNGRLCGVFVNISLYGFGIAYIITASISIR 601
Query: 153 AIERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYST 212
AI+ S CFH + C + M+ FG +++VLSQIP+F + WLSI+AA+MSF Y+
Sbjct: 602 AIQDSICFHNKGSEKTCGYDDTYNMLGFGAIQVVLSQIPNFHNIEWLSIVAAIMSFAYAF 781
Query: 213 IGLGLGFGKVIENG 226
IG+GL ENG
Sbjct: 782 IGMGLAIV*GHENG 823
>AL379703 homologue to GP|6539600|gb| amino acid transporter a {Vicia faba},
partial (60%)
Length = 481
Score = 188 bits (478), Expect = 3e-48
Identities = 96/160 (60%), Positives = 122/160 (76%), Gaps = 2/160 (1%)
Frame = +1
Query: 114 DVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKSEGKDPCHMNG 173
+VV S +GG + +LCG+ QY+NL GV IGYTI +SISM+A++RSNC+HK C+++
Sbjct: 1 EVVRSVLGGRKFQLCGLAQYINLIGVTIGYTITASISMVAVKRSNCYHKQGHDAKCYISN 180
Query: 174 NIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKVIENG-AVGGSL 232
N +MI F ++IVLSQIP+F +L WLSI+AAVMSF YS+IGLGL KV G AV SL
Sbjct: 181 NPFMIIFACIQIVLSQIPNFHKLSWLSIVAAVMSFAYSSIGLGLSIAKVAGRGPAVRTSL 360
Query: 233 TGITVGT-VTQTQKVWRTLQALGDIAFAYSYSMILIEIQD 271
TG+ VG VT T+KVWR QA+GDIAFAY+YS +LIEIQD
Sbjct: 361 TGVQVGVDVTGTEKVWRMFQAIGDIAFAYAYSNVLIEIQD 480
>TC82881 similar to GP|15216026|emb|CAC51423. amino acid permease AAP1
{Vicia faba var. minor}, partial (28%)
Length = 1132
Score = 181 bits (458), Expect(2) = 7e-47
Identities = 88/140 (62%), Positives = 105/140 (74%), Gaps = 5/140 (3%)
Frame = +2
Query: 10 KNNHHHQAFDVSLD-----MQQQGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSL 64
K+ H+ FD+ ++ + SK +DDD RLKRTGTVWT S+HIITAV+GSGVLSL
Sbjct: 641 KDLEHNLEFDMEVEHSIEAVPSHKDSKLYDDDDRLKRTGTVWTTSSHIITAVVGSGVLSL 820
Query: 65 AWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQ 124
AWAIAQLGWV GP+VMI FSL+T+YTS LL CYR GDP GKRNYT+M+ H+ +GG
Sbjct: 821 AWAIAQLGWVIGPSVMIFFSLITWYTSSLLAECYRIGDPHYGKRNYTFMEAGHTILGGFN 1000
Query: 125 VKLCGIVQYLNLFGVAIGYT 144
LCGIVQY NL+G AIGYT
Sbjct: 1001DTLCGIVQYTNLYGTAIGYT 1060
Score = 25.0 bits (53), Expect(2) = 7e-47
Identities = 10/15 (66%), Positives = 14/15 (92%)
Frame = +1
Query: 146 ASSISMIAIERSNCF 160
A +ISM+AI+RS+CF
Sbjct: 1066 AGAISMMAIKRSDCF 1110
>TC82248 similar to GP|15216028|emb|CAC51424. amino acid permease AAP3
{Vicia faba var. minor}, partial (29%)
Length = 779
Score = 174 bits (440), Expect = 9e-44
Identities = 82/122 (67%), Positives = 97/122 (79%)
Frame = +3
Query: 27 QGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLV 86
Q SK +DDDGR+KRTGT+WT +HIITAVIGSGVLSLAW+IAQ+GWVAGP MI FS++
Sbjct: 414 QDDSKYYDDDGRVKRTGTIWTTCSHIITAVIGSGVLSLAWSIAQMGWVAGPGAMIFFSII 593
Query: 87 TYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIA 146
T YTS L CYR GD GKRNYT+MD V + +GG VK+CGIVQYLNLFG A+ Y IA
Sbjct: 594 TLYTSSFLADCYRCGDTEFGKRNYTFMDAVSNILGGPSVKICGIVQYLNLFGSAMRYNIA 773
Query: 147 SS 148
++
Sbjct: 774 AA 779
>BQ147947 weakly similar to GP|10176848|db amino acid transporter
{Arabidopsis thaliana}, partial (24%)
Length = 521
Score = 148 bits (374), Expect = 4e-36
Identities = 64/137 (46%), Positives = 95/137 (68%), Gaps = 2/137 (1%)
Frame = -3
Query: 337 IVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQI--PLFNAYKINLFRLVWRTI 394
IV+H VG YQ+YS P + + + + K+P+S FV Q+ PL A++ N+ R+ +RT
Sbjct: 519 IVLHXVGGYQIYSXPXYTAADRWCSRKYPNSGFVNNFYQLKLPLLPAFQXNMLRICFRTA 340
Query: 395 FVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSL 454
+VI TT ++++ P+FN+++G+LGALGFWPL +YFPVEMY +Q +I W KWI + FS
Sbjct: 339 YVISTTGLAIMFPYFNEVLGVLGALGFWPLXIYFPVEMYFVQNKIEAWXTKWIVXRTFSF 160
Query: 455 ACLIVTVAAAAGSVAGI 471
CL+VTV + GS+ GI
Sbjct: 159 VCLLVTVVSLVGSLEGI 109
>TC92351 homologue to GP|6539604|gb|AAF15946.1| amino acid transporter c
{Vicia faba}, partial (28%)
Length = 497
Score = 122 bits (307), Expect = 2e-28
Identities = 59/103 (57%), Positives = 71/103 (68%)
Frame = +1
Query: 19 DVSLDMQQQGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPA 78
D +Q S +DDDG KRTG + +A AHIITAVIGSGVLSLAW+ AQLGW+ GP
Sbjct: 91 DGKSSLQITKASGAYDDDGHAKRTGNLKSAVAHIITAVIGSGVLSLAWSTAQLGWIGGPV 270
Query: 79 VMILFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMG 121
++ ++VTY S LL CYRN D V GKRNY+YMD V N+G
Sbjct: 271 TLLCCAIVTYIPSFLLSDCYRNPDSVTGKRNYSYMDAVRVNLG 399
>AW736648 similar to PIR|T50691|T506 amino acid permease 6 [imported] -
Arabidopsis thaliana, partial (19%)
Length = 493
Score = 120 bits (300), Expect = 2e-27
Identities = 53/96 (55%), Positives = 75/96 (77%)
Frame = -3
Query: 388 RLVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWI 447
R+V RT +V+IT LI+M+ PFFND +GL+G+L FWPLTVYFP+EMYI Q ++ ++S W
Sbjct: 491 RVV*RTAYVVITALIAMIFPFFNDFLGLIGSLSFWPLTVYFPIEMYIKQSKMQRFSFTWT 312
Query: 448 CFQLFSLACLIVTVAAAAGSVAGIFLDLQVYKPFKA 483
++ S ACLIV++ +AAGS+ G+ DL+ Y+PFKA
Sbjct: 311 WMKILSWACLIVSIISAAGSIQGLAHDLKKYQPFKA 204
>AL384207 weakly similar to GP|21553710|gb| amino acid carrier putative
{Arabidopsis thaliana}, partial (17%)
Length = 517
Score = 115 bits (287), Expect = 5e-26
Identities = 50/99 (50%), Positives = 73/99 (73%)
Frame = +2
Query: 383 KINLFRLVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKW 442
K+NLFRL+WRTIFVI+ T+++M +PFFN + LLGA+GF PL V+FP++M+I QKRIP
Sbjct: 2 KLNLFRLIWRTIFVILATILAMAMPFFNQFLALLGAIGFGPLVVFFPIQMHIAQKRIPVL 181
Query: 443 SIKWICFQLFSLACLIVTVAAAAGSVAGIFLDLQVYKPF 481
S++W QL + C++V++AA S+ I ++ YK F
Sbjct: 182 SLRWCALQLLNCLCMVVSLAAIVASIHEISENIHKYKIF 298
>TC78342 similar to GP|8571474|gb|AAF76897.1| proline/glycine betaine
transporter {Atriplex hortensis}, partial (90%)
Length = 1717
Score = 95.1 bits (235), Expect = 5e-20
Identities = 122/471 (25%), Positives = 193/471 (40%), Gaps = 32/471 (6%)
Frame = +3
Query: 46 WTASAHIITAVIGSG-VLSLAWAI-AQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDP 103
W A ++T + S VL + I LGWV G ++L + ++ Y + L+ + G
Sbjct: 231 WFQVAFVLTTGVNSAYVLGYSGTIMVPLGWVGGVVGLVLAAAISLYANALIAMLHEYG-- 404
Query: 104 VNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKS 163
G R+ Y D+ G + +QY+NLF + GY I + A++ + +
Sbjct: 405 --GTRHIRYRDLAGYIYGKKAYSITWTLQYINLFMINTGYIILAGS---ALKAAYTVFRD 569
Query: 164 EG--KDP-CHMNGNIYMISFGLVEIVLSQIPDFDQLW-WLSILAAVMSFTYSTIGLGLGF 219
+G K P C I F + IP L WL + + V +F Y I L L
Sbjct: 570 DGVLKLPYCIAIAGIVCAMFAIC------IPHLSALGVWLGV-STVFTFVYIVIALVLSI 728
Query: 220 GKVIENGAVGGSLT--GITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPP 277
+ + A ++ G+T K++ T+ A + FAY+ M L EIQ T+K P
Sbjct: 729 KDGMNSPARDYAVPEHGVT--------KIFTTIGASASLVFAYNTGM-LPEIQATIKQPV 881
Query: 278 SESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAI 337
K M K+ + + + + GY A+G+ + LL P W+ +AN
Sbjct: 882 V--KNMMKSLWFQFTIGLVPMYMVTFAGYWAYGNKTETYLLNSVN--GPAWVKALANITA 1049
Query: 338 VIHLVGAYQVYSQPLFAFVEN----------------YAAEKF--PDSDFVTKTVQIPLF 379
+ V A +Y+ P++ + + A + F P +++ I
Sbjct: 1050FLQSVIALHIYANPIYEYFDTD*GIVLNQLKLNNMSLIA*QIFASPMYEYLDTRFGISGE 1229
Query: 380 NAYKINL-FRLVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKR 438
NL FR+ R ++ T I+ LLPF D L GA+ +PLT MY K+
Sbjct: 1230AMKAKNLSFRVGVRGGYLAFNTFIAALLPFLGDFESLTGAISTFPLTFILANHMYYKAKK 1409
Query: 439 IPKWSIK-----WICFQLFSLACLIVTVAAAAGSVAGIFLDLQVYKPFKAI 484
K SI W FSL + TVAA + I +D + Y F I
Sbjct: 1410-NKLSISQKGGLWANIVFFSLMSIAATVAA----IRLIAVDSKTYSLFADI 1547
>BQ152966 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsis thaliana},
partial (31%)
Length = 574
Score = 73.2 bits (178), Expect = 2e-13
Identities = 46/167 (27%), Positives = 76/167 (44%), Gaps = 11/167 (6%)
Frame = +2
Query: 329 LIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFR 388
LI AN +VIH++G+YQ+Y+ P+F +E +K N + R
Sbjct: 2 LIATANMFVVIHVIGSYQIYAMPVFDMIETLLVKKM---------------NFEPSTMLR 136
Query: 389 LVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPK----WSI 444
+ R ++V T I++ PFF+ ++G G F P T + P M++ + K W
Sbjct: 137 FIVRNVYVAFTMFIAITFPFFDGLLGFFGGFAFAPTTYFLPCIMWLSIYKPRKFGLSWWA 316
Query: 445 KWICFQLFSLACLIVTVAAAAGSVAGIFLDLQVYK-------PFKAI 484
WIC L CL+ + + G + I + + YK PFK++
Sbjct: 317 NWICIVLG--VCLM--ILSPIGGLRTIIIKAKTYKFYS*TPSPFKSV 445
>TC77442 similar to GP|20804928|dbj|BAB92607. proline transport protein-like
{Oryza sativa (japonica cultivar-group)}, partial (62%)
Length = 1795
Score = 70.9 bits (172), Expect = 1e-12
Identities = 96/415 (23%), Positives = 167/415 (40%), Gaps = 4/415 (0%)
Frame = +3
Query: 71 LGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGI 130
LGW G ++ + L T Y + LL A + ++G+R Y D++ G L
Sbjct: 486 LGWTWGIILLFVIGLYTAYANWLLAAFHF----IDGRRFIRYRDLMGFVYGTKMYHLTWT 653
Query: 131 VQYLNLFGVAIGYTIASSISMIAIERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLS-Q 189
Q+L L +G+ + ++ I F S P + Y++ G + S
Sbjct: 654 SQFLTLLLGNMGFILLGGKALKEINAE--FSDS----PWRLQ--YYIVVTGAAYFIFSFS 809
Query: 190 IPDFDQLW-WLSILAAV-MSFTYSTIGLGLGFGKVIENGAVGGSLTGITVGTVTQTQKVW 247
IP + WL A V +++ + + + GK N S++G ++ KV+
Sbjct: 810 IPTLSSMRNWLGASAVVTLAYIAFLVSVAVKDGK--SNSDKDYSVSG------SKVNKVF 965
Query: 248 RTLQALGDIAFAYSYSMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYA 307
+ A+ I + M L EIQ T++ P K M+KA + V LFY GY
Sbjct: 966 NSFGAISAIIVTNTSGM-LPEIQSTLRKPAV--KNMRKALYSQYTVGALFYYGVTIVGYW 1136
Query: 308 AFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDS 367
A+G L P W+ + N + + + ++ P+ ++
Sbjct: 1137AYGSMVSSYLPENLS--GPRWINVLVNVIVFLQSAVSQHLFVVPIHEALD---------- 1280
Query: 368 DFVTKTVQIPLFNAYKINLFRL-VWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTV 426
T+ ++I NL RL + R F T I+ PF D V LLG+ PLT
Sbjct: 1281---TRFLEIGKGMHSGENLKRLFLLRMCFYTGNTFIAAAFPFMGDFVNLLGSFSLVPLTF 1451
Query: 427 YFPVEMYIIQKRIPKWSIKWICFQLFSLACLIVTVAAAAGSVAGIFLDLQVYKPF 481
FP +++ K + K + + + ++T+A ++ I ++Q Y+ F
Sbjct: 1452MFPSMIFLKIKGKTARTEKKVWHWINIVVSFLLTIATTISALRFIINNVQKYQFF 1616
>BF633909 similar to GP|13676299|gb| amino acid transporter {Glycine max},
partial (11%)
Length = 328
Score = 69.3 bits (168), Expect = 3e-12
Identities = 32/34 (94%), Positives = 33/34 (96%)
Frame = +2
Query: 30 SKCFDDDGRLKRTGTVWTASAHIITAVIGSGVLS 63
SKCFDDDGRLKRTGT WTA+AHIITAVIGSGVLS
Sbjct: 227 SKCFDDDGRLKRTGTFWTATAHIITAVIGSGVLS 328
>CB892086 similar to GP|6539604|gb|A amino acid transporter c {Vicia faba},
partial (18%)
Length = 462
Score = 65.9 bits (159), Expect = 3e-11
Identities = 32/55 (58%), Positives = 40/55 (72%)
Frame = +2
Query: 169 CHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKVI 223
C IYMI FGLV++++S IPD + LS++AAVMSFTYS+IGLGLG VI
Sbjct: 11 CKYGDTIYMILFGLVQVIMSFIPDLHNMALLSVVAAVMSFTYSSIGLGLGVTNVI 175
>TC79749 weakly similar to GP|15290010|dbj|BAB63704. contains ESTs
C99526(E20351) C73742(E20351)~unknown protein {Oryza
sativa (japonica cultivar-group)}, partial (36%)
Length = 655
Score = 58.2 bits (139), Expect = 7e-09
Identities = 37/157 (23%), Positives = 80/157 (50%), Gaps = 1/157 (0%)
Frame = +2
Query: 51 HIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNY 110
H+ T+++G +L+L ++ LGW G ++L ++T+Y+ LL + + G+R +
Sbjct: 209 HLTTSIVGPVILTLPFSFTLLGWFGGVIWLVLAGVITFYSYNLLSIVLEHHAQL-GRRQF 385
Query: 111 TYMDVVHSNMGGIQVK-LCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKSEGKDPC 169
+ D+ +G K G +Q++ FG IG + S+ I + + EG
Sbjct: 386 RFRDMARDILGPRWAKYYIGPLQFIICFGTVIGGPLVGGKSLKFIYQ---LYHPEGSMKL 556
Query: 170 HMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVM 206
+ ++I G+V ++L+Q+P F L +++++ ++
Sbjct: 557 YQ----FIIICGVVTMLLAQLPSFHSLRHINLISLIL 655
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.141 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,974,858
Number of Sequences: 36976
Number of extensions: 254937
Number of successful extensions: 1980
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 1936
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1957
length of query: 485
length of database: 9,014,727
effective HSP length: 100
effective length of query: 385
effective length of database: 5,317,127
effective search space: 2047093895
effective search space used: 2047093895
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0181.18


