

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0180a.6
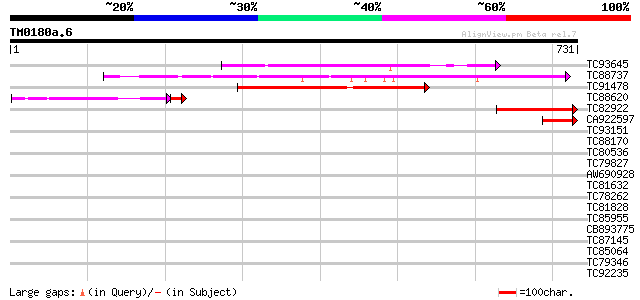
(731 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC93645 similar to GP|19699053|gb|AAL90894.1 AT5g23390/T32G24_2 ... 280 1e-75
TC88737 similar to PIR|A96737|A96737 hypothetical protein F3I17.... 256 2e-68
TC91478 similar to GP|15810447|gb|AAL07111.1 unknown protein {Ar... 199 3e-51
TC88620 weakly similar to GP|15810447|gb|AAL07111.1 unknown prot... 109 4e-27
TC82922 weakly similar to PIR|B96526|B96526 unknown protein [imp... 104 1e-22
CA922597 91 1e-18
TC93151 homologue to GP|22136928|gb|AAM91808.1 unknown protein {... 32 0.87
TC88170 weakly similar to GP|17104695|gb|AAL34236.1 unknown prot... 32 1.1
TC80536 similar to GP|6520227|dbj|BAA87955.1 ZCW7 {Arabidopsis t... 31 1.9
TC79827 weakly similar to PIR|T46136|T46136 remorin-like protein... 31 1.9
AW690928 similar to GP|22093675|d putative pre-mRNA splicing fac... 30 2.5
TC81632 similar to GP|20279471|gb|AAM18751.1 unknown protein {Or... 30 3.3
TC78262 similar to GP|20260620|gb|AAM13208.1 unknown protein {Ar... 30 4.3
TC81828 similar to GP|9758905|dbj|BAB09481.1 contains similarity... 29 5.6
TC85955 similar to GP|3860319|emb|CAA10127.1 nucleolar protein {... 29 5.6
CB893775 similar to GP|8978074|dbj protein serine/threonine kina... 29 7.4
TC87145 similar to PIR|E71446|E71446 probable protein kinase - A... 29 7.4
TC85064 similar to GP|15529208|gb|AAK97698.1 At2g01710/T8O11.12 ... 29 7.4
TC79346 similar to GP|9294216|dbj|BAB02118.1 contains similarity... 29 7.4
TC92235 weakly similar to GP|15029315|gb|AAK81821.1 CYFIP2 {Mus ... 29 7.4
>TC93645 similar to GP|19699053|gb|AAL90894.1 AT5g23390/T32G24_2
{Arabidopsis thaliana}, partial (26%)
Length = 994
Score = 280 bits (717), Expect = 1e-75
Identities = 177/366 (48%), Positives = 213/366 (57%), Gaps = 7/366 (1%)
Frame = +3
Query: 274 GISAWPGRLTLTNYALYFESLGVGVHEKPVRYDLGTDLKQVIKPDLTGPLGARLFDKAVM 333
GI+AWPGRLTLTN L + K + + PL K +
Sbjct: 3 GIAAWPGRLTLTNMLCTLSHLALVCMRKLFGMIWAQT*NRS*SLN*LDPLVLVYLIK--L 176
Query: 334 YKSTSVAEPVYFEFPEFKANVRRDYWLDISLEILRVHKFIRKYYLKETQKAEVLARAILG 393
++ + PVYFEFPEFKAN+RRDYWLDISLEILR H+F+RK+ LK+TQK EVLARA LG
Sbjct: 177 *CTSQLL*PVYFEFPEFKANLRRDYWLDISLEILRAHRFVRKFGLKDTQKLEVLARANLG 356
Query: 394 IYRYRAVREAFKYFSSHYKTLLSFNLAEALPRGDMILETLSNSLTNLTVASSKRSSPRNV 453
I+RYRA++EAFK+FSS+YKTLL+FNLAE LPRGD IL+TLSNSLTN T AS KR NV
Sbjct: 357 IFRYRALKEAFKFFSSNYKTLLTFNLAETLPRGDRILQTLSNSLTNSTAASGKRDISANV 536
Query: 454 DTKKQLVVSPASAVALFRLGFKSDKFADIYEETTVV------GDIRVGE-INLLEMAVKK 506
+ KKQ VSPA+ VALF LGFKS K ADIYEETT D +G + K
Sbjct: 537 ERKKQSAVSPAAVVALFCLGFKSKKAADIYEETTACL*YPSW*DKSLGHGSKTIPSGHWK 716
Query: 507 SLRDTGKAEAAQATVDQVKVEGIDTNVAVMKELLFPVIESANRLERLASWKDFYKSTAFL 566
S TG +++ D+ K + + P +T F
Sbjct: 717 SRSCTGNCGPSESGGDRYKCCSYEGTTIPSHRIC*P-------------------TTTFS 839
Query: 567 LLTSYMILRGWIHYLLPSIFVVVAILMLWRRHFRKGRSLEAFIVTPPPNRNPVEQLLTLQ 626
L+ + +AI+MLWRRHFRKG +LE F V PPPNRN VEQLLTLQ
Sbjct: 840 LVERLLF---------------IAIIMLWRRHFRKGGALEPFTVRPPPNRNAVEQLLTLQ 974
Query: 627 EAITQF 632
EAITQF
Sbjct: 975 EAITQF 992
>TC88737 similar to PIR|A96737|A96737 hypothetical protein F3I17.11
[imported] - Arabidopsis thaliana, partial (62%)
Length = 2115
Score = 256 bits (655), Expect = 2e-68
Identities = 183/632 (28%), Positives = 310/632 (48%), Gaps = 30/632 (4%)
Frame = +1
Query: 121 LADKDFRRLTYDMMLAWETPSVHTDETPPPSSKDEHGGDDDEASLFYSSSTNMAVQVDDQ 180
L D F+RL + MLAWE P Y S+N
Sbjct: 37 LQDPAFQRLIFITMLAWENPYT------------------------YVLSSNAEKASLQS 144
Query: 181 KTVGLEAFSRIAPVCAPIADVITVHNLFDALTSSSGRRLHFLVYDKYIRFLDKVIKNSKN 240
K V EAF RIAP + + D TVHNLF L + ++ ++++ +K +
Sbjct: 145 KRVTEEAFVRIAPAVSGVVDRPTVHNLFKVLAGDKDG----ISMSTWLAYINEFVKVRRE 312
Query: 241 VLASSVGSLQLAEEEIVLDVDGTIPTQPVLQHIGISAWPGRLTLTNYALYFESLGVGVHE 300
+ + +EE +L + G+ QPVL+ AWPG+LTLT+ A+YFE G+ ++
Sbjct: 313 NRSYQIPEFPQIDEEKILCI-GSNSKQPVLKWENNMAWPGKLTLTDKAIYFEGAGLLGNK 489
Query: 301 KPVRYDLGTDLKQVIKPDLTGPLGARLFDKAVMYKSTSVAEPVYFEFPEFKANVRRDYWL 360
+ +R DL D +V K + GPLG+ LFD AV S S + EF + ++RRD W
Sbjct: 490 RAMRLDLTYDGLRVEKAKV-GPLGSSLFDSAVSISSGSESNWWVLEFIDLGGDMRRDVWH 666
Query: 361 DISLEILRVHKFIRKY---------YLKETQKAEVLARAILGIYRYRAVREAFKYFSSHY 411
+ E++ +HKF +Y + K + AI GI R +A++ K
Sbjct: 667 ALISEVIALHKFTHEYGPDEYGPNVFEARKGKQRATSSAINGIARLQALQHLRKLLDDPT 846
Query: 412 KTLLSFNLAEALPRGDMILETLSNSLTN---LTVASSKRSSPRNVDTKK----QLVVSPA 464
K L+ F+ + P GD++L++L+ + +T +S R P N + + V
Sbjct: 847 K-LVQFSYLQNAPNGDIVLQSLAVNYWGSQLVTGFTSTRHQPENRPSNEIADSSNHVFDI 1023
Query: 465 SAVALFRLGFKSDKFAD-----IYEETTVVGDIR-----VGEINLLEMAVKKSLRDTGKA 514
R KS + ++ T+ G + V +++L E A K S + +
Sbjct: 1024DGSVYLRKWMKSPSWGSSTSTSFWKNTSTKGLVLSKNHVVADLSLTERAAKTSKQKSQVV 1203
Query: 515 EAAQATVDQVKVEGIDTNVAVMKELLFPVIESANRLERLASWKDFYKSTAFLLLTSYMIL 574
E QAT+D ++GI +N+ + KEL+FP+ +A E+L W++ + + FL L +I
Sbjct: 1204EKTQATIDAATLKGIPSNIDLFKELIFPITLTAKNFEKLRHWEEPHLTVGFLGLAYTLIF 1383
Query: 575 RGWIHYLLPSIFVVVAILMLWRRHFRK----GRSLEAFIVTPPPNRNPVEQLLTLQEAIT 630
R + Y+ P + ++ A+ ML R ++ GR ++ P N +++++ +++A+
Sbjct: 1384RNLLSYIFPVMLMITAVGMLTIRSLKEQGRLGRFFGGVMIRDQPPSNTIQKIIAVKDAMR 1563
Query: 631 QFESLIQAGNIVLLKVRALLLALLPQATEKVALLLVFIAAVFAFVPPKYIIFVIFFEYYT 690
E++ Q N+ LLK+R++LL+ PQ T +VA+L++ A + VP KYI+ + F+ +T
Sbjct: 1564DVENMTQKVNVSLLKIRSILLSGNPQITTEVAVLMLTWATILFIVPFKYILSFLLFDMFT 1743
Query: 691 REMPCRKESSDRWVRRMREWWIRIPAAPVELI 722
RE+ R+E +R + +RE W +PAAPV ++
Sbjct: 1744RELEFRREMVERLTKLLRERWHAVPAAPVAVL 1839
>TC91478 similar to GP|15810447|gb|AAL07111.1 unknown protein {Arabidopsis
thaliana}, partial (27%)
Length = 736
Score = 199 bits (506), Expect = 3e-51
Identities = 118/250 (47%), Positives = 155/250 (61%), Gaps = 2/250 (0%)
Frame = +3
Query: 294 LGVGVHEKPVRYDLGTDLKQVIKPDLTGPLGARLFDKAVMYKSTSVAEPVYFEFPEFKAN 353
L V ++KP RYDL DL QV+KP+LTGP G RLFDKAV Y STS++EP EFPE K +
Sbjct: 6 LRVVSYDKPKRYDLADDLNQVVKPELTGPWGTRLFDKAVFYSSTSLSEPAVLEFPELKGH 185
Query: 354 VRRDYWLDISLEILRVHKFIRKYYLKETQKAEVLARAILGIYRYRAVREAFKYFSSHYKT 413
RRDYWL I EIL VHK+I K+ +K + E L +A+LGI R +A+++ +
Sbjct: 186 ARRDYWLAIIREILCVHKYISKFGIKGVARDEALWKAVLGILRLQAIQDISSLGPVPNDS 365
Query: 414 LLSFNLAEALPRGDMILETLSNSLTNLTVASSKRSSPRNVDTKKQLVVSPASAV-ALFRL 472
LL FNL + LP GD+ILETL N S+ S D+K + + S + + L
Sbjct: 366 LLMFNLCDQLPGGDLILETLVN-------MSNSSESNHGHDSKPESGMYSISVLDTVSNL 524
Query: 473 GFKSDKFADIYEETTV-VGDIRVGEINLLEMAVKKSLRDTGKAEAAQATVDQVKVEGIDT 531
GF ++ E+ + VG+I VGE LLE VK+S + K +AQAT D VKV+GIDT
Sbjct: 525 GFVFGTSSNSSVESRIAVGEITVGETTLLERVVKESKNNYKKVVSAQATSDGVKVDGIDT 704
Query: 532 NVAVMKELLF 541
N+AVMKELL+
Sbjct: 705 NLAVMKELLY 734
>TC88620 weakly similar to GP|15810447|gb|AAL07111.1 unknown protein
{Arabidopsis thaliana}, partial (25%)
Length = 785
Score = 109 bits (272), Expect(2) = 4e-27
Identities = 76/206 (36%), Positives = 106/206 (50%)
Frame = +2
Query: 3 ENGSENKSGGMWENFLRNHQNSLKSLFLRNSSSSKNKTDTTADDSAVNSPKPIPLLSPLA 62
E + K+ M+E +++ SLK L + S ++ + SA + LSP+A
Sbjct: 194 EMATAGKTRAMFEGLVKD--GSLKWLLGKKSYFAEEFEEMENSPSA--GKNFVRELSPVA 361
Query: 63 NSVVVRSSKILGISTEELQHAFDSELPLGVKELLTYARNLLEFCSYKALHKLSKSSDYLA 122
N VV R +KIL S+ +LQ F+ E +K YARNLLE+C +KAL ++ S +L
Sbjct: 362 NLVVRRCAKILKTSSSDLQEGFNLEASDSLKNPSRYARNLLEYCCFKALSLKAQISGHLL 541
Query: 123 DKDFRRLTYDMMLAWETPSVHTDETPPPSSKDEHGGDDDEASLFYSSSTNMAVQVDDQKT 182
DK FRRLTYDMMLAWE P ++++ VD++ +
Sbjct: 542 DKTFRRLTYDMMLAWEVP---------------------------AATSQPLTNVDEEVS 640
Query: 183 VGLEAFSRIAPVCAPIADVITVHNLF 208
VGLEAF RIAP IA+VI NLF
Sbjct: 641 VGLEAFCRIAPAVPIIANVIICENLF 718
Score = 30.8 bits (68), Expect(2) = 4e-27
Identities = 12/21 (57%), Positives = 17/21 (80%)
Frame = +3
Query: 208 FDALTSSSGRRLHFLVYDKYI 228
F+ L+SS+G RL F +YDKY+
Sbjct: 717 FEVLSSSTGGRLQFPIYDKYL 779
>TC82922 weakly similar to PIR|B96526|B96526 unknown protein [imported] -
Arabidopsis thaliana, partial (12%)
Length = 648
Score = 104 bits (260), Expect = 1e-22
Identities = 51/104 (49%), Positives = 68/104 (65%)
Frame = +1
Query: 628 AITQFESLIQAGNIVLLKVRALLLALLPQATEKVALLLVFIAAVFAFVPPKYIIFVIFFE 687
A++Q E I GN+ LLK R L L+ PQAT+K+A L+ V AF+P KYI+ ++F
Sbjct: 1 AVSQAEQFILDGNLFLLKFRGLWLSFFPQATDKMAFGLLSAGLVLAFLPTKYIVLLVFLN 180
Query: 688 YYTREMPCRKESSDRWVRRMREWWIRIPAAPVELIKPDECKKRK 731
+T P RK S++RW RR REWW IPAAPV+L + E KK+K
Sbjct: 181 TFTMFSPPRKASTERWERRFREWWFSIPAAPVKLERDKEDKKKK 312
>CA922597
Length = 750
Score = 91.3 bits (225), Expect = 1e-18
Identities = 38/45 (84%), Positives = 42/45 (92%)
Frame = -2
Query: 687 EYYTREMPCRKESSDRWVRRMREWWIRIPAAPVELIKPDECKKRK 731
E YTREMPCR+ESS RW+RR REWWIRIPAAPVEL+KP+ECKKRK
Sbjct: 749 ECYTREMPCRRESSKRWIRRFREWWIRIPAAPVELVKPEECKKRK 615
>TC93151 homologue to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (4%)
Length = 657
Score = 32.0 bits (71), Expect = 0.87
Identities = 23/69 (33%), Positives = 35/69 (50%), Gaps = 3/69 (4%)
Frame = +1
Query: 54 PIPLLSPLANSVVVRSSKILGISTEE---LQHAFDSELPLGVKELLTYARNLLEFCSYKA 110
P +L PLA +VV ++ GIS E LQH + ++E L + LL+ CS +
Sbjct: 340 PFSVLPPLARRLVVELPQLRGISLAELHRLQHRY-------LEERLRVQQRLLQQCSEER 498
Query: 111 LHKLSKSSD 119
+L + SD
Sbjct: 499 HRQLLRRSD 525
>TC88170 weakly similar to GP|17104695|gb|AAL34236.1 unknown protein
{Arabidopsis thaliana}, partial (95%)
Length = 1146
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 2/65 (3%)
Frame = +2
Query: 26 KSLFLRNSSSSKNKT--DTTADDSAVNSPKPIPLLSPLANSVVVRSSKILGISTEELQHA 83
K+LF ++ S S + T D ++DD ++N+ P S A VVVR + + ++Q
Sbjct: 89 KALFTKSLSHSHSITMDDFSSDDESINNNSSSPTPSDYAEIVVVRHGETAWNAISKVQGQ 268
Query: 84 FDSEL 88
D EL
Sbjct: 269 LDVEL 283
>TC80536 similar to GP|6520227|dbj|BAA87955.1 ZCW7 {Arabidopsis thaliana},
partial (55%)
Length = 1022
Score = 30.8 bits (68), Expect = 1.9
Identities = 17/48 (35%), Positives = 26/48 (53%)
Frame = -1
Query: 13 MWENFLRNHQNSLKSLFLRNSSSSKNKTDTTADDSAVNSPKPIPLLSP 60
++EN + N F RNSS SK T++D+++ P PIP+ P
Sbjct: 644 LFENGMSNKHRR----FRRNSSLSKPPRRNTSNDNSIRLPLPIPIPIP 513
>TC79827 weakly similar to PIR|T46136|T46136 remorin-like protein -
Arabidopsis thaliana, partial (71%)
Length = 776
Score = 30.8 bits (68), Expect = 1.9
Identities = 14/49 (28%), Positives = 28/49 (56%)
Frame = +1
Query: 514 AEAAQATVDQVKVEGIDTNVAVMKELLFPVIESANRLERLASWKDFYKS 562
++ A ++VDQ G DT +V ++ + +ES RL + +W++ K+
Sbjct: 157 SDHATSSVDQTTAAGTDTKDSVDRDAVLARVESQKRLALIKAWEENEKT 303
>AW690928 similar to GP|22093675|d putative pre-mRNA splicing factor
ATP-dependent RNA helicase {Oryza sativa (japonica
cultivar-group), partial (10%)
Length = 638
Score = 30.4 bits (67), Expect = 2.5
Identities = 28/112 (25%), Positives = 49/112 (43%), Gaps = 6/112 (5%)
Frame = +1
Query: 19 RNHQNSLKSLFLRNSSSSKNKTDTTADDSAVNSPKPIPLLSPLANSVVVRS------SKI 72
R HQ S +F+ S ++ + S+ NSP P +SP + V +R+ S +
Sbjct: 109 RRHQPSPSPMFVGASPDARLVSPWHTPHSSYNSPSPWDHVSP--SPVPIRASGSSVKSSV 282
Query: 73 LGISTEELQHAFDSELPLGVKELLTYARNLLEFCSYKALHKLSKSSDYLADK 124
G + + AF SE +E + +L E Y+ + + +Y AD+
Sbjct: 283 SGYNRRSHKLAFSSENSDTYEEEIADKSDLGEEHKYEITESMRQEMEYDADR 438
>TC81632 similar to GP|20279471|gb|AAM18751.1 unknown protein {Oryza sativa
(japonica cultivar-group)}, partial (32%)
Length = 821
Score = 30.0 bits (66), Expect = 3.3
Identities = 18/50 (36%), Positives = 24/50 (48%), Gaps = 5/50 (10%)
Frame = -3
Query: 140 PSVHTDETPPPSS-----KDEHGGDDDEASLFYSSSTNMAVQVDDQKTVG 184
P VH T P +DE GGDDDEA ++ + N ++D VG
Sbjct: 318 PQVHKGNTEKPKQDPYEDEDEDGGDDDEA*VWVVDN-NQGNDIEDNAWVG 172
>TC78262 similar to GP|20260620|gb|AAM13208.1 unknown protein {Arabidopsis
thaliana}, partial (93%)
Length = 3014
Score = 29.6 bits (65), Expect = 4.3
Identities = 18/63 (28%), Positives = 28/63 (43%), Gaps = 4/63 (6%)
Frame = +3
Query: 554 ASWKDFYKSTAFLLLTSYMILRGWIHYLLPS----IFVVVAILMLWRRHFRKGRSLEAFI 609
A W D + LL S ++L G + + +V+ IL +W + KGR AF+
Sbjct: 2142 AFWPDVHFRVIIALLVSQIVLMGLLTTKKAASSTPFLIVLPILTIWFHRYCKGRFESAFV 2321
Query: 610 VTP 612
P
Sbjct: 2322 KFP 2330
>TC81828 similar to GP|9758905|dbj|BAB09481.1 contains similarity to
transcription regulator~gene_id:MRG7.19 {Arabidopsis
thaliana}, partial (10%)
Length = 1298
Score = 29.3 bits (64), Expect = 5.6
Identities = 19/60 (31%), Positives = 30/60 (49%)
Frame = +3
Query: 537 KELLFPVIESANRLERLASWKDFYKSTAFLLLTSYMILRGWIHYLLPSIFVVVAILMLWR 596
K L V+E A L+ L + + LLL+++ +L +H LLPS V + + WR
Sbjct: 222 KPHLLKVVELALLLQHLLA------TMLLLLLSTFQVLICPVHQLLPSFLVQILFEISWR 383
>TC85955 similar to GP|3860319|emb|CAA10127.1 nucleolar protein {Cicer
arietinum}, partial (98%)
Length = 1981
Score = 29.3 bits (64), Expect = 5.6
Identities = 18/57 (31%), Positives = 29/57 (50%)
Frame = -1
Query: 617 NPVEQLLTLQEAITQFESLIQAGNIVLLKVRALLLALLPQATEKVALLLVFIAAVFA 673
NPV+ L A+T ++ + I LL++ L+ +LP T K A LV + F+
Sbjct: 586 NPVDIKFNLSSAVTVAKTKLCLFQISLLEITNKLVKMLPDTTNKFAHKLVTLNRNFS 416
>CB893775 similar to GP|8978074|dbj protein serine/threonine kinase-like
{Arabidopsis thaliana}, partial (50%)
Length = 858
Score = 28.9 bits (63), Expect = 7.4
Identities = 20/54 (37%), Positives = 27/54 (49%)
Frame = +1
Query: 87 ELPLGVKELLTYARNLLEFCSYKALHKLSKSSDYLADKDFRRLTYDMMLAWETP 140
E+ LG E L Y LE + +++ KSS+ L D+DF D LA E P
Sbjct: 511 EIMLGAAEGLAYLHEGLEI---QVIYRDFKSSNVLLDEDFHPKLSDFGLAREGP 663
>TC87145 similar to PIR|E71446|E71446 probable protein kinase - Arabidopsis
thaliana, partial (74%)
Length = 1443
Score = 28.9 bits (63), Expect = 7.4
Identities = 20/54 (37%), Positives = 27/54 (49%)
Frame = +2
Query: 87 ELPLGVKELLTYARNLLEFCSYKALHKLSKSSDYLADKDFRRLTYDMMLAWETP 140
E+ LG E L Y LE + +++ KSS+ L D+DF D LA E P
Sbjct: 752 EIMLGAAEGLAYLHEGLEI---QVIYRDFKSSNVLLDEDFHPKLSDFGLAREGP 904
>TC85064 similar to GP|15529208|gb|AAK97698.1 At2g01710/T8O11.12
{Arabidopsis thaliana}, partial (29%)
Length = 645
Score = 28.9 bits (63), Expect = 7.4
Identities = 13/31 (41%), Positives = 18/31 (57%)
Frame = +3
Query: 599 FRKGRSLEAFIVTPPPNRNPVEQLLTLQEAI 629
FRK R + +V PPNR+P+ + QE I
Sbjct: 261 FRKARQQQP*LVLHPPNRSPIRRPRPYQETI 353
>TC79346 similar to GP|9294216|dbj|BAB02118.1 contains similarity to unknown
protein~gb|AAB61017.1~gene_id:K17E12.5 {Arabidopsis
thaliana}, partial (96%)
Length = 1707
Score = 28.9 bits (63), Expect = 7.4
Identities = 16/36 (44%), Positives = 23/36 (63%)
Frame = +3
Query: 24 SLKSLFLRNSSSSKNKTDTTADDSAVNSPKPIPLLS 59
+L +L+ SSSSKN T T ++S +N K I L+S
Sbjct: 201 NLFALYAFTSSSSKNHTTTNTNNSNLNLHKNISLVS 308
>TC92235 weakly similar to GP|15029315|gb|AAK81821.1 CYFIP2 {Mus musculus},
partial (2%)
Length = 628
Score = 28.9 bits (63), Expect = 7.4
Identities = 12/44 (27%), Positives = 23/44 (52%)
Frame = +1
Query: 124 KDFRRLTYDMMLAWETPSVHTDETPPPSSKDEHGGDDDEASLFY 167
KD R+ DM +T+++ +HGG++ +A++FY
Sbjct: 22 KDLSRILSDMRTLSADWMANTNKSESELQSSQHGGEESKANIFY 153
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,930,617
Number of Sequences: 36976
Number of extensions: 288696
Number of successful extensions: 1724
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 1688
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1716
length of query: 731
length of database: 9,014,727
effective HSP length: 103
effective length of query: 628
effective length of database: 5,206,199
effective search space: 3269492972
effective search space used: 3269492972
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0180a.6


