

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
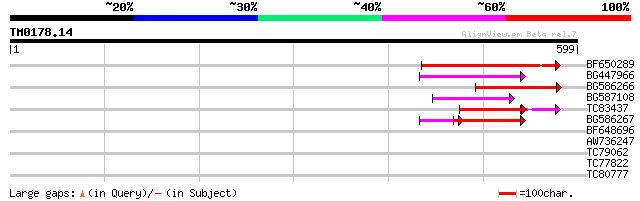
Query= TM0178.14
(599 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 122 5e-28
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 86 5e-17
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 76 4e-14
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 71 1e-12
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 55 3e-11
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 59 7e-09
BF648696 31 1.5
AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase... 31 1.5
TC79062 homologue to GP|13344808|gb|AAK19054.1 UVI1 {Pisum sativ... 30 2.6
TC77822 weakly similar to GP|21536834|gb|AAM61166.1 unknown {Ara... 29 4.5
TC80777 homologue to GP|4115386|gb|AAD03387.1| unknown protein {... 28 7.7
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 122 bits (305), Expect = 5e-28
Identities = 59/147 (40%), Positives = 95/147 (64%)
Frame = +3
Query: 436 KFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITH 495
K G DG N F+K W+ + + +A FF TG +P IN T VTL+PK N S+ +
Sbjct: 72 KAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFMPKIINCTYVTLLPKEVNVTSVKN 251
Query: 496 YRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILRK 555
+RPI+CC+ YK+IS+I+ +R++G LN +++ NQSAF+ ++I DNI+++ E + RK
Sbjct: 252 FRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVKGRVIFDNIILSHELVKSYSRK 431
Query: 556 GRASKEHVALKLDMSKAYERVEWSFLE 582
G + + +K+D+ KAY+ EW F++
Sbjct: 432 GISPR--CMVKIDLXKAYDSXEWPFIK 506
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 85.5 bits (210), Expect = 5e-17
Identities = 42/112 (37%), Positives = 64/112 (56%)
Frame = +2
Query: 434 PRKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESI 493
P K GPDGL LF++ +W V + + + ++N+T + LIPK NP +
Sbjct: 299 PVKAPGPDGLPALFFQKYWHIVGKEVQQMVLQVLNNSMETEELNKTFIVLIPKGKNPNTP 478
Query: 494 THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIA 545
YRPIS CN K+I+++I R+K L +I + QSAF+ ++I DN +IA
Sbjct: 479 KDYRPISLCNVVMKIITKVIANRVKQTLPDVIDVEQSAFVQGRLITDNALIA 634
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 75.9 bits (185), Expect = 4e-14
Identities = 33/91 (36%), Positives = 59/91 (64%)
Frame = -3
Query: 493 ITHYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNI 552
++ YR I+ CN YK+I++I+ R++ L +I+ +QSAF+ + I DN++I + H +
Sbjct: 778 VSEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYL 599
Query: 553 LRKGRASKEHVALKLDMSKAYERVEWSFLEK 583
+ G +A+K DM+KAY+R+ W+FL +
Sbjct: 598 RQSGAKKHVSMAVKTDMTKAYDRIAWNFLRE 506
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 71.2 bits (173), Expect = 1e-12
Identities = 34/87 (39%), Positives = 52/87 (59%)
Frame = +3
Query: 447 FYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITHYRPISCCNFTY 506
F+++ W + + SF ++G L +++N T + LIPK P +T RPIS CN Y
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 507 KVISRIIVTRLKGGLNQLITLNQSAFI 533
K+IS+++ RLK L LI+ QSAF+
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFV 671
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 54.7 bits (130), Expect(2) = 3e-11
Identities = 25/71 (35%), Positives = 43/71 (60%)
Frame = +2
Query: 476 INETVVTLIPKVPNPESITHYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGV 535
IN T + LIPKV NP+ + +RPIS YK++ +++ RL+ + +I+ QSAF+
Sbjct: 50 INSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVVIGSVISDAQSAFVKN 229
Query: 536 KMIQDNIMIAQ 546
+ I + + + Q
Sbjct: 230 RQILEMVFL*Q 262
Score = 31.6 bits (70), Expect(2) = 3e-11
Identities = 15/43 (34%), Positives = 25/43 (57%)
Frame = +1
Query: 540 DNIMIAQEAFHNILRKGRASKEHVALKLDMSKAYERVEWSFLE 582
D I+IA EA + + K+ + K+D KAY V+W++L+
Sbjct: 244 DGILIANEA---VDEAKKLKKDLLLFKVDFEKAYHSVDWAYLD 363
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 58.5 bits (140), Expect = 7e-09
Identities = 29/76 (38%), Positives = 46/76 (60%)
Frame = +1
Query: 470 GHLPSKINETVVTLIPKVPNPESITHYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQ 529
G+L + + L+PK + + +RPIS CN YK++S+++ RLK L +IT Q
Sbjct: 562 GNLKRESTKQTSGLVPKKLEAKRLVEFRPISLCNVAYKIVSKVLSKRLKSVLPWIITETQ 741
Query: 530 SAFIGVKMIQDNIMIA 545
+AF ++I DNI+IA
Sbjct: 742 AAFGRRQLISDNILIA 789
Score = 44.3 bits (103), Expect = 1e-04
Identities = 19/46 (41%), Positives = 26/46 (56%)
Frame = +3
Query: 434 PRKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINET 479
P K GPDG+N F++ WDT+ + A+ F TG L IN+T
Sbjct: 453 PHKCPGPDGMNVFFFQQFWDTMGDDLTSMAQEFLRTGKLEEGINKT 590
>BF648696
Length = 653
Score = 30.8 bits (68), Expect = 1.5
Identities = 19/53 (35%), Positives = 24/53 (44%), Gaps = 6/53 (11%)
Frame = +2
Query: 253 VQLIQEVPGKSTWPEAKHAKTISS------LGIRAPS*QRTKRSPSLNRSWPF 299
VQ +Q + G TWPEA + I S + AP T P +N WPF
Sbjct: 230 VQKVQVIIGMHTWPEAAIMEDIGSKAQVPIISFAAP----TITPPLMNNRWPF 376
>AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (4%)
Length = 305
Score = 30.8 bits (68), Expect = 1.5
Identities = 12/34 (35%), Positives = 17/34 (49%)
Frame = +1
Query: 439 GPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHL 472
GPDG+N F K +W+ + F F + G L
Sbjct: 184 GPDGVNFTFVKEYWELIKVDFLRVVMEFHTNGKL 285
>TC79062 homologue to GP|13344808|gb|AAK19054.1 UVI1 {Pisum sativum},
partial (79%)
Length = 567
Score = 30.0 bits (66), Expect = 2.6
Identities = 12/31 (38%), Positives = 17/31 (54%)
Frame = +3
Query: 33 WRPPRQ*ES*KTYPKKKDRQSCFLWRQELRG 63
W P Q E + ++D+ SC WRQE+ G
Sbjct: 111 WFQPIQKEEQQRQSPRQDQDSCIFWRQEVEG 203
>TC77822 weakly similar to GP|21536834|gb|AAM61166.1 unknown {Arabidopsis
thaliana}, partial (33%)
Length = 2391
Score = 29.3 bits (64), Expect = 4.5
Identities = 14/41 (34%), Positives = 22/41 (53%)
Frame = +1
Query: 493 ITHYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFI 533
+TH P C F + + +++RLK N ++TL S FI
Sbjct: 76 VTHNPPFHCSLFLSLL*ALFLLSRLKNKRNTIVTLPSSLFI 198
>TC80777 homologue to GP|4115386|gb|AAD03387.1| unknown protein {Arabidopsis
thaliana}, partial (39%)
Length = 1307
Score = 28.5 bits (62), Expect = 7.7
Identities = 15/43 (34%), Positives = 20/43 (45%)
Frame = +1
Query: 464 RSFFSTGHLPSKINETVVTLIPKVPNPESITHYRPISCCNFTY 506
R F+T +LP LI +P+ +YR CCNF Y
Sbjct: 1165 RGTFTTSYLPRLWQMLTFVLISALPS----FYYRSYFCCNFLY 1281
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.350 0.155 0.548
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,474,264
Number of Sequences: 36976
Number of extensions: 278983
Number of successful extensions: 2419
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 1340
Number of HSP's successfully gapped in prelim test: 70
Number of HSP's that attempted gapping in prelim test: 1046
Number of HSP's gapped (non-prelim): 1457
length of query: 599
length of database: 9,014,727
effective HSP length: 102
effective length of query: 497
effective length of database: 5,243,175
effective search space: 2605857975
effective search space used: 2605857975
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0178.14


