

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0177.1
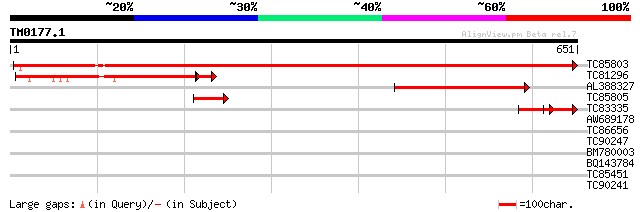
(651 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85803 similar to GP|13785471|dbj|BAB43909. phosphoenolpyruvate... 1165 0.0
TC81296 similar to GP|6707571|gb|AAF25671.1| ATP-dependent phosp... 291 7e-85
AL388327 similar to GP|15983749|gb phosphoenolpyruvate carboxyki... 225 5e-59
TC85805 weakly similar to SP|P35334|PGIP_PHAVU Polygalacturonase... 75 6e-14
TC83335 similar to GP|13785471|dbj|BAB43909. phosphoenolpyruvate... 56 5e-08
AW689178 homologue to PIR|T06779|T0 clathrin heavy chain - soybe... 35 0.12
TC86656 similar to GP|7108521|gb|AAF36454.1| ubiquitin-protein l... 32 0.77
TC90247 weakly similar to GP|21592898|gb|AAM64848.1 unknown {Ara... 31 1.3
BM780003 weakly similar to PIR|T06165|T0 multidrug resistance pr... 30 2.2
BQ143784 weakly similar to SP|P33485|VNUA Probable nuclear antig... 29 5.0
TC85451 similar to PIR|T06239|T06239 probable glutathione transf... 29 5.0
TC90241 similar to GP|15724302|gb|AAL06544.1 AT5g64830/MXK3_5 {A... 29 6.5
>TC85803 similar to GP|13785471|dbj|BAB43909. phosphoenolpyruvate
carboxykinase {Flaveria pringlei}, partial (95%)
Length = 2351
Score = 1165 bits (3013), Expect = 0.0
Identities = 580/657 (88%), Positives = 608/657 (92%), Gaps = 10/657 (1%)
Frame = +3
Query: 5 NGNG-IAT----NGLPSIHTQK--NGICHDDSVPTVKANTIDELHSLQKKKSAPGTPISG 57
NGNG IAT NGLP I T+K NGICHDDS PTV A TIDELHSLQKKKSAP TP +G
Sbjct: 153 NGNGSIATENGSNGLPKITTKKSVNGICHDDSGPTVNAKTIDELHSLQKKKSAPSTPNTG 332
Query: 58 TQTPFT--SDPERQQQQLQSISASLASLTRETGPKVVKGDPAKKLENPKTIHQVSHHHI- 114
T TPF SD ERQ+ QLQSISASLASLTRETGPKVVKGDPAK N K H V HHH+
Sbjct: 333 TSTPFAQISDQERQKLQLQSISASLASLTRETGPKVVKGDPAK---NQKVDH-VHHHHVP 500
Query: 115 APTIAVSDSALKFTHVLYNLSPAELYEQAIRYEKGSFVTSTGALATLSGAKTGRSPRDKR 174
PTIAVSDSALKFTHVLYNLSPAELYEQAI+YEKGSF+TS GA+ATLSGAKTGRSPRDKR
Sbjct: 501 TPTIAVSDSALKFTHVLYNLSPAELYEQAIKYEKGSFITSNGAMATLSGAKTGRSPRDKR 680
Query: 175 VVRDAVTEDDLWWGKGSPNIEMDEQTFLVNRERAVDYLNSLDKVFVNDQFLNWDPENRIK 234
VV+D VTE++LWWGKGSPNIEMDE+TF+VNRERAVDYLNSLDKVFVNDQFLNWDPENRIK
Sbjct: 681 VVKDKVTENELWWGKGSPNIEMDEETFMVNRERAVDYLNSLDKVFVNDQFLNWDPENRIK 860
Query: 235 VRIVAARAYHSLFMHNMCIRPTPEELENFGTPDFTIYNAGQFPCNRFTHYMTSSTSVDLN 294
VRIV+ARAYHSLFMHNMCIRP+PEELENFGTPDFTIYNAG+FPCNRFTHYMTSSTS+DLN
Sbjct: 861 VRIVSARAYHSLFMHNMCIRPSPEELENFGTPDFTIYNAGKFPCNRFTHYMTSSTSIDLN 1040
Query: 295 LARREMVILGTQYAGEMKKGLFGVMHYLMPKRKILSLHSGCNMGKDGDVALFFGLSGTGK 354
LARREMVILGTQYAGEMKKGLF VMHYLMPKR+ILSLHSG NMGK GDVALFFGLSGTGK
Sbjct: 1041LARREMVILGTQYAGEMKKGLFSVMHYLMPKRQILSLHSGSNMGKGGDVALFFGLSGTGK 1220
Query: 355 TTLSTDHNRYLIGDDEHCWSDKGVSNIEGGCYAKCIDLSREKEPDIWNAIRFGTVLENVV 414
TTLSTDHNRYLIGDDEHCWS+ GVSNIEGGCYAKCIDLSREKEPDIWNAIRFGTVLENVV
Sbjct: 1221TTLSTDHNRYLIGDDEHCWSENGVSNIEGGCYAKCIDLSREKEPDIWNAIRFGTVLENVV 1400
Query: 415 FDEHTREVDYSDKSVTENTRAAYPIEYIPNAKLPCVGPHPTNVILLACDAFGVLPPVSKL 474
FDEHTREVDYSDKSVTENTRAAYPIEYIPNAKLPCVGPHP NVILLACDAFGVLPPVSKL
Sbjct: 1401FDEHTREVDYSDKSVTENTRAAYPIEYIPNAKLPCVGPHPKNVILLACDAFGVLPPVSKL 1580
Query: 475 NLAQTMYHFISGYTALVAGTEDGIKEPQATFSACFGAAFIMLHPTKYAAMLAEKMEQHGA 534
LAQTMYHFISGYTALVAGTEDGIKEPQATFSACFGAAFIMLHPTKYAAMLAEKME HGA
Sbjct: 1581TLAQTMYHFISGYTALVAGTEDGIKEPQATFSACFGAAFIMLHPTKYAAMLAEKMESHGA 1760
Query: 535 TGWLVNTGWSGGSYGSGSRIKLQYTRKIIDAIHSGSLLKAEFQKTPIFGLEIPTEVEGVP 594
TGWLVNTGWSGGSYG+G+RIKL YTRKIIDAIH+GSLL AE++K+ IFGL+ PTEVEGVP
Sbjct: 1761TGWLVNTGWSGGSYGTGNRIKLSYTRKIIDAIHNGSLLGAEYKKSEIFGLQTPTEVEGVP 1940
Query: 595 SEILDPVNTWSDKDAYQETLLKLAGLFKNNFETFTNYKIGKGDNNLTEEILAAGPNF 651
SEILDP+N WSDK+AY TLLKL GLFK NFETFTNYKIGKGD+NLTEEILAAGP F
Sbjct: 1941SEILDPINAWSDKNAYNATLLKLGGLFKKNFETFTNYKIGKGDDNLTEEILAAGPIF 2111
>TC81296 similar to GP|6707571|gb|AAF25671.1| ATP-dependent
phosphoenolpyruvate carboxykinase {Medicago sativa},
partial (44%)
Length = 813
Score = 291 bits (744), Expect(2) = 7e-85
Identities = 160/232 (68%), Positives = 178/232 (75%), Gaps = 19/232 (8%)
Frame = +3
Query: 7 NGIATNGLPSIHTQK-----NGICHDDSVPTVKANTIDELHSLQKKK---SAPGTPISG- 57
N + GLP I T K + ICHDDS P+VKA TI ELHSLQKKK S P TPI
Sbjct: 66 NVVGRRGLPMIQTHKKVMDSSEICHDDSSPSVKAQTIHELHSLQKKKNNKSTPNTPIRSS 245
Query: 58 --TQTPFTS----DPERQQQQLQSISASLASLTRETGPKVVKGDPAKKLENPKTIHQVSH 111
TQ+PF S + ++Q+QQLQSIS SLASLTRETGPKVVKGDPA K + V H
Sbjct: 246 TTTQSPFASISEEERQKQKQQLQSISESLASLTRETGPKVVKGDPASKGSSS----HVDH 413
Query: 112 HHIAPTIA----VSDSALKFTHVLYNLSPAELYEQAIRYEKGSFVTSTGALATLSGAKTG 167
H +P I+ VSDSALKFTHVLYNLSPAELYEQAI+YEKGSF+T+TGALATLSGAKTG
Sbjct: 414 HQHSPLISTAFDVSDSALKFTHVLYNLSPAELYEQAIKYEKGSFITATGALATLSGAKTG 593
Query: 168 RSPRDKRVVRDAVTEDDLWWGKGSPNIEMDEQTFLVNRERAVDYLNSLDKVF 219
RSPRDKRVV+D T+ DLWWGKGSPNIEMDE TFLVNRERAVDYLNSL+KV+
Sbjct: 594 RSPRDKRVVKDQTTQHDLWWGKGSPNIEMDEHTFLVNRERAVDYLNSLEKVY 749
Score = 42.4 bits (98), Expect(2) = 7e-85
Identities = 18/23 (78%), Positives = 20/23 (86%)
Frame = +1
Query: 215 LDKVFVNDQFLNWDPENRIKVRI 237
L + VNDQFLNWDPE+RIKVRI
Sbjct: 736 LKRFIVNDQFLNWDPEHRIKVRI 804
>AL388327 similar to GP|15983749|gb phosphoenolpyruvate carboxykinase
{Emericella nidulans}, partial (28%)
Length = 481
Score = 225 bits (573), Expect = 5e-59
Identities = 102/157 (64%), Positives = 126/157 (79%), Gaps = 1/157 (0%)
Frame = +3
Query: 442 IPNAKLPCVGP-HPTNVILLACDAFGVLPPVSKLNLAQTMYHFISGYTALVAGTEDGIKE 500
IPNAK+PC+ HP N+ILL CDA+G+LPPVSKL+ +Q MYHFISGYTA +AGTE+G+ E
Sbjct: 3 IPNAKIPCISSDHPKNIILLTCDAYGILPPVSKLSSSQAMYHFISGYTAKIAGTEEGVTE 182
Query: 501 PQATFSACFGAAFIMLHPTKYAAMLAEKMEQHGATGWLVNTGWSGGSYGSGSRIKLQYTR 560
P+ATFSACFG F++LHP +YA MLA+KME H A WL+NTGW+GG+YG G RIKL+Y+R
Sbjct: 183 PEATFSACFGQPFLVLHPYRYAKMLADKMEHHKADAWLINTGWNGGAYGVGERIKLKYSR 362
Query: 561 KIIDAIHSGSLLKAEFQKTPIFGLEIPTEVEGVPSEI 597
IIDAIHSG L KAE++ I L+IP GVP+EI
Sbjct: 363 AIIDAIHSGELTKAEYENYDILNLQIPKSCSGVPTEI 473
>TC85805 weakly similar to SP|P35334|PGIP_PHAVU Polygalacturonase inhibitor
precursor (Polygalacturonase-inhibiting protein).,
partial (26%)
Length = 922
Score = 75.5 bits (184), Expect = 6e-14
Identities = 34/40 (85%), Positives = 37/40 (92%)
Frame = -1
Query: 212 LNSLDKVFVNDQFLNWDPENRIKVRIVAARAYHSLFMHNM 251
++S VFVNDQFLNWDPENRIKVRIV+ARAYHSLFMHNM
Sbjct: 142 ISSFVSVFVNDQFLNWDPENRIKVRIVSARAYHSLFMHNM 23
>TC83335 similar to GP|13785471|dbj|BAB43909. phosphoenolpyruvate
carboxykinase {Flaveria pringlei}, partial (9%)
Length = 535
Score = 55.8 bits (133), Expect = 5e-08
Identities = 28/38 (73%), Positives = 32/38 (83%)
Frame = +3
Query: 614 LLKLAGLFKNNFETFTNYKIGKGDNNLTEEILAAGPNF 651
LLKLAGLFKNNF+ F +YKIGK D LTEEI++AGP F
Sbjct: 90 LLKLAGLFKNNFDGFVSYKIGK-DQKLTEEIVSAGPIF 200
Score = 53.1 bits (126), Expect = 3e-07
Identities = 22/41 (53%), Positives = 31/41 (74%)
Frame = +2
Query: 585 EIPTEVEGVPSEILDPVNTWSDKDAYQETLLKLAGLFKNNF 625
EIPT ++GVPSEILDP+NTW +K AYQ+T +++ + F
Sbjct: 2 EIPTAIDGVPSEILDPINTWFEKKAYQDTPSEISWSIQEQF 124
>AW689178 homologue to PIR|T06779|T0 clathrin heavy chain - soybean, partial
(10%)
Length = 545
Score = 34.7 bits (78), Expect = 0.12
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 5/105 (4%)
Frame = +3
Query: 392 LSREKEPDIW-----NAIRFGTVLENVVFDEHTREVDYSDKSVTENTRAAYPIEYIPNAK 446
LS ++PDI A + G + E + TRE + D T+N ++ AK
Sbjct: 60 LSSSEDPDIHFKYIEAAAKTGQIKE---VERVTRESSFYDPEKTKN--------FLMEAK 206
Query: 447 LPCVGPHPTNVILLACDAFGVLPPVSKLNLAQTMYHFISGYTALV 491
LP P ++ CD FG +P ++ M +I GY V
Sbjct: 207 LPDARP-----LINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKV 326
>TC86656 similar to GP|7108521|gb|AAF36454.1| ubiquitin-protein ligase 1
{Arabidopsis thaliana}, partial (12%)
Length = 1939
Score = 32.0 bits (71), Expect = 0.77
Identities = 23/110 (20%), Positives = 39/110 (34%), Gaps = 6/110 (5%)
Frame = +3
Query: 17 IHTQKNGICHDDSVPTVKANTIDELHSLQKKKSAPGTPISGTQTPFTSDPERQQQQLQSI 76
+H + G HD S+P + +K S P + F E+ ++ L +
Sbjct: 360 LHPAQPGASHDQSIPVISDVENASTSESPQKVSGPAVKVDEKNMAFVKFSEKHRKLLNAF 539
Query: 77 SASLASLTRET------GPKVVKGDPAKKLENPKTIHQVSHHHIAPTIAV 120
L ++ P+ + D + K HQ HHH I+V
Sbjct: 540 IRQNPGLLEKSFLLMLKVPRFIDFDNKRAHFRSKIKHQHDHHHSPLRISV 689
>TC90247 weakly similar to GP|21592898|gb|AAM64848.1 unknown {Arabidopsis
thaliana}, partial (60%)
Length = 1041
Score = 31.2 bits (69), Expect = 1.3
Identities = 31/144 (21%), Positives = 54/144 (36%), Gaps = 5/144 (3%)
Frame = +1
Query: 360 DHNRYLIGDDEHCWSDKGVSNIEGGCYAKCIDLSREKEPDIWNAIRFGTVLENVVFDEHT 419
+ N Y EH V GGC A + S ++ +W+ R GT+L ++VF
Sbjct: 565 EKNLYEYSFSEHTMRVTDVVIGNGGCNAIIVSASDDRTCKVWSLSR-GTLLRSIVFPS-- 735
Query: 420 REVDYSDKSVTENTRAAYPIEYI-----PNAKLPCVGPHPTNVILLACDAFGVLPPVSKL 474
N P E++ + K+ + + + ++ S
Sbjct: 736 ----------VINAIVLDPAEHVFYAGSSDGKIFIAALNTERITTIDNYGMHIIGSFSNH 885
Query: 475 NLAQTMYHFISGYTALVAGTEDGI 498
+ A T + G L++G+EDGI
Sbjct: 886 SKAVTCLAYSKGRNLLISGSEDGI 957
>BM780003 weakly similar to PIR|T06165|T0 multidrug resistance protein 1
homolog - barley, partial (12%)
Length = 665
Score = 30.4 bits (67), Expect = 2.2
Identities = 23/70 (32%), Positives = 31/70 (43%), Gaps = 6/70 (8%)
Frame = +2
Query: 58 TQTPFTSDPERQQQQLQSISASLA------SLTRETGPKVVKGDPAKKLENPKTIHQVSH 111
TQ D ++ Q Q QSI S A + T + P ++ P L N I QVSH
Sbjct: 143 TQLSINDDQDQNQNQEQSILLSAARSSAGRTSTARSSPLIL---PKSPLPNDIIISQVSH 313
Query: 112 HHIAPTIAVS 121
H P+ + S
Sbjct: 314 SHSHPSPSFS 343
>BQ143784 weakly similar to SP|P33485|VNUA Probable nuclear antigen. [strain
Kaplan PRV] {Pseudorabies virus}, partial (2%)
Length = 1022
Score = 29.3 bits (64), Expect = 5.0
Identities = 13/42 (30%), Positives = 22/42 (51%)
Frame = +2
Query: 31 PTVKANTIDELHSLQKKKSAPGTPISGTQTPFTSDPERQQQQ 72
P + LH+ +KK+ PGTP + +TP P ++ Q+
Sbjct: 230 PKTANQKANPLHASPQKKTHPGTPATPRETPSRRTPAQRGQR 355
>TC85451 similar to PIR|T06239|T06239 probable glutathione transferase (EC
2.5.1.18) 2 4-D inducible - soybean, complete
Length = 1155
Score = 29.3 bits (64), Expect = 5.0
Identities = 20/68 (29%), Positives = 31/68 (45%), Gaps = 17/68 (25%)
Frame = -1
Query: 106 IHQVSHHHIAPTI--AVSDSALKFTHVLYNLSPAELYEQAIR---------------YEK 148
+H+ S+ H P I +SD K T + + P + EQ ++ YE+
Sbjct: 474 LHRSSYQHNQPRI*LVISDKDQKATKEISHSKPHQYTEQQLKIHKLIYHYE*EQGFSYEQ 295
Query: 149 GSFVTSTG 156
GSFVT+ G
Sbjct: 294 GSFVTTKG 271
>TC90241 similar to GP|15724302|gb|AAL06544.1 AT5g64830/MXK3_5 {Arabidopsis
thaliana}, partial (18%)
Length = 952
Score = 28.9 bits (63), Expect = 6.5
Identities = 17/48 (35%), Positives = 25/48 (51%)
Frame = +2
Query: 435 AAYPIEYIPNAKLPCVGPHPTNVILLACDAFGVLPPVSKLNLAQTMYH 482
A YPI + ++ +GP P +L C F L PV+ L+L + M H
Sbjct: 251 ANYPIPLLLKSEASLIGPFPRIPSILIC-FFVPLVPVASLSLHKFMLH 391
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,604,407
Number of Sequences: 36976
Number of extensions: 264580
Number of successful extensions: 1425
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1414
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1419
length of query: 651
length of database: 9,014,727
effective HSP length: 102
effective length of query: 549
effective length of database: 5,243,175
effective search space: 2878503075
effective search space used: 2878503075
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0177.1


