

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
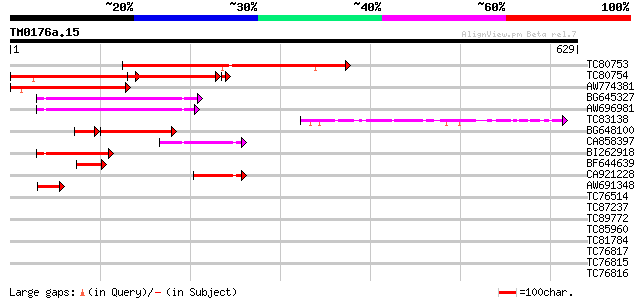
Query= TM0176a.15
(629 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80753 homologue to PIR|T06153|T06153 hypothetical protein F24J... 348 4e-96
TC80754 homologue to GP|9759604|dbj|BAB11392.1 contains similari... 233 1e-61
AW774381 homologue to GP|9759604|dbj contains similarity to cycl... 211 9e-55
BG645327 similar to GP|13924511|gb ania-6a type cyclin {Arabidop... 114 1e-25
AW696981 similar to GP|13924511|gb ania-6a type cyclin {Arabidop... 110 2e-24
TC83138 similar to GP|9759604|dbj|BAB11392.1 contains similarity... 97 2e-20
BG648100 74 2e-13
CA858397 similar to GP|11340587|em unnamed protein product {Zea ... 69 5e-12
BI262918 similar to GP|13365979|dbj putative cyclin homolog {Ory... 67 2e-11
BF644639 similar to GP|9759604|dbj| contains similarity to cycli... 62 7e-10
CA921228 57 2e-08
AW691348 weakly similar to GP|12744989|gb| putative cyclin {Arab... 42 5e-04
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 37 0.030
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 37 0.030
TC89772 similar to GP|9759529|dbj|BAB10995.1 gene_id:K21L19.3~un... 35 0.087
TC85960 similar to GP|23094138|gb|AAF50778.2 CG4835-PA {Drosophi... 35 0.087
TC81784 similar to PIR|T40294|T40294 hypothetical protein SPBC35... 31 1.3
TC76817 GP|2598577|emb|CAA75576.1 MtN22 {Medicago truncatula}, c... 31 1.6
TC76815 MtN22 31 1.6
TC76816 GP|2598577|emb|CAA75576.1 MtN22 {Medicago truncatula}, p... 31 1.6
>TC80753 homologue to PIR|T06153|T06153 hypothetical protein F24J7.161 -
Arabidopsis thaliana, partial (27%)
Length = 900
Score = 348 bits (893), Expect = 4e-96
Identities = 193/265 (72%), Positives = 208/265 (77%), Gaps = 12/265 (4%)
Frame = +1
Query: 126 VSYEIIHKKDPAAVQRIKQKEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKK 185
VSYEII+KKDP AVQRIKQKEVYEQQKELILL ERVVLATLGFDF+V HPYKPLVEAIKK
Sbjct: 1 VSYEIINKKDPTAVQRIKQKEVYEQQKELILLAERVVLATLGFDFNVHHPYKPLVEAIKK 180
Query: 186 LKVSQNALAQVAWNFVNDGLRTSLCLQFKPQHIAAGAIFLAAKFLNVKL----EKLWWQV 241
KV+QNALAQVAWNFVNDGLRTSLCLQFKP HIAAGAIFLAAKFL VKL EK+WWQ
Sbjct: 181 FKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQE 360
Query: 242 FDVTLTAHQLEEVSNQMLELYEQNQIAPSNDVEGTAGGGN--RATPKASTSNDEAATTNN 299
FDV T QLEEVSNQMLELYEQN++ PSN+ EGT GG R T KA TSNDE A N+
Sbjct: 361 FDV--TPRQLEEVSNQMLELYEQNRMPPSNEAEGTTGGAASIRTTVKAPTSNDETAPANS 534
Query: 300 NLYIRGPSPRLEASKLAISKNVV--SSANHVG-PVSNHGTN---GSTEMIHLVEGDAKGN 353
N G S RLE SK A SK S+AN VG PVSN G + GSTE+ H VE D KGN
Sbjct: 535 NSQTGGISSRLETSKPASSKTAFDSSAANQVGRPVSNPGRSRDYGSTEIKHRVEDDVKGN 714
Query: 354 QHPEQEPPPHKENLQEAQDTVRSRS 378
QHPE EP P+K+NLQ+ Q+ VRSRS
Sbjct: 715 QHPEHEPLPYKDNLQQVQNVVRSRS 789
>TC80754 homologue to GP|9759604|dbj|BAB11392.1 contains similarity to
cyclin~gene_id:K18C1.7 {Arabidopsis thaliana}, partial
(38%)
Length = 1115
Score = 233 bits (595), Expect = 1e-61
Identities = 119/149 (79%), Positives = 129/149 (85%), Gaps = 4/149 (2%)
Frame = +2
Query: 1 MAGLLLGDTSHDGTHQSGSQGCSQE---NGSRWYLSRKEIEENSPSKQDGVDLKKEAYLR 57
MAGLLLGD SH G H GSQ S+E +GSRWY SRKEIEENSPS++DG+DLKKE YLR
Sbjct: 203 MAGLLLGDASHQGAHPVGSQRYSEEKPEDGSRWYFSRKEIEENSPSQEDGIDLKKETYLR 382
Query: 58 KSYCTYLQDLGMRLKVPQVTIASAIIFCHRFFLRQSHAKNDRRIIATSCMFLAGKVEETP 117
KSYCT+LQDLGMRLKVPQVTIA+AIIFCHRFFLRQSHAKNDRR IAT CMFLAGKVEETP
Sbjct: 383 KSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRTIATVCMFLAGKVEETP 562
Query: 118 RPLKDVILVSYEIIHK-KDPAAVQRIKQK 145
RPLKDVI+VSYEII+K + QRIKQK
Sbjct: 563 RPLKDVIMVSYEIINKERIQQQFQRIKQK 649
Score = 168 bits (425), Expect(2) = 2e-42
Identities = 85/104 (81%), Positives = 91/104 (86%)
Frame = +3
Query: 131 IHKKDPAAVQRIKQKEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQ 190
I K ++ + +KEVYEQQKELILL ERVVLATLGFDF+V HPYKPLVEAIKK KV+Q
Sbjct: 606 IKKGSNSSFRESNRKEVYEQQKELILLAERVVLATLGFDFNVHHPYKPLVEAIKKFKVAQ 785
Query: 191 NALAQVAWNFVNDGLRTSLCLQFKPQHIAAGAIFLAAKFLNVKL 234
NALAQVAWNFVNDGLRTSLCLQFKP HIAAGAIFLAAKFL VKL
Sbjct: 786 NALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKL 917
Score = 23.1 bits (48), Expect(2) = 2e-42
Identities = 7/10 (70%), Positives = 9/10 (90%)
Frame = +2
Query: 236 KLWWQVFDVT 245
++WWQ FDVT
Sbjct: 932 EVWWQEFDVT 961
>AW774381 homologue to GP|9759604|dbj contains similarity to
cyclin~gene_id:K18C1.7 {Arabidopsis thaliana}, partial
(18%)
Length = 628
Score = 211 bits (536), Expect = 9e-55
Identities = 104/138 (75%), Positives = 117/138 (84%), Gaps = 4/138 (2%)
Frame = +1
Query: 1 MAGLLLGDTSH----DGTHQSGSQGCSQENGSRWYLSRKEIEENSPSKQDGVDLKKEAYL 56
MA L+ G+ H DG+ SQ +E RWY+SRKEIEEN+PS++DG+DLKKE YL
Sbjct: 214 MAALVSGELPHHGASDGSSCRSSQDKQEEALGRWYMSRKEIEENAPSRKDGIDLKKETYL 393
Query: 57 RKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFFLRQSHAKNDRRIIATSCMFLAGKVEET 116
RKSYCT+LQDLGMRLKVPQVTIA+AIIFCHRFFLRQSHAKNDRR IAT CMFLAGKVEET
Sbjct: 394 RKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRTIATVCMFLAGKVEET 573
Query: 117 PRPLKDVILVSYEIIHKK 134
PRPLKDVIL+SYE+IHKK
Sbjct: 574 PRPLKDVILISYEMIHKK 627
>BG645327 similar to GP|13924511|gb ania-6a type cyclin {Arabidopsis
thaliana}, partial (51%)
Length = 789
Score = 114 bits (285), Expect = 1e-25
Identities = 60/186 (32%), Positives = 111/186 (59%), Gaps = 1/186 (0%)
Frame = +1
Query: 30 WYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFF 89
+YL+ +++ NSPS++DG+D E LR C +Q+ G+ L++PQ +A+ + HRF+
Sbjct: 172 FYLTDEQLT-NSPSRKDGIDEATETTLRIYGCDLIQESGILLRLPQAVMATGQVLFHRFY 348
Query: 90 LRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIK-QKEVY 148
++S A+ + + +A SC++LA K+EE R + V+++ + + +++ + + + Y
Sbjct: 349 CKKSFARFNVKKVAASCVWLASKLEENTRKARQVLIIFHRMECRRENLPIDHLDLYSKKY 528
Query: 149 EQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVNDGLRTS 208
K + ER +L +GF HV+HP+K + + L+ L+Q AWN ND LRT+
Sbjct: 529 VDLKTELSRTERHILKEMGFICHVEHPHKFISNYLATLETPPE-LSQEAWNLANDSLRTT 705
Query: 209 LCLQFK 214
LC++F+
Sbjct: 706 LCVRFR 723
>AW696981 similar to GP|13924511|gb ania-6a type cyclin {Arabidopsis
thaliana}, partial (45%)
Length = 648
Score = 110 bits (275), Expect = 2e-24
Identities = 59/182 (32%), Positives = 107/182 (58%), Gaps = 1/182 (0%)
Frame = +3
Query: 30 WYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFF 89
+YL+ +++ NSPS++DG+D E LR C +Q+ G+ L++PQ +A+ + HRF+
Sbjct: 108 FYLTDEQLT-NSPSRKDGIDEATETTLRIYGCDLIQESGILLRLPQAVMATGQVLFHRFY 284
Query: 90 LRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIK-QKEVY 148
++S A+ + + +A SC++LA K+EE R + V+++ + + +++ + + + Y
Sbjct: 285 CKKSFARFNVKKVAASCVWLASKLEENTRKARQVLIIFHRMECRRENLPIDHLDLYSKKY 464
Query: 149 EQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVNDGLRTS 208
K + ER +L +GF HV+HP+K + + L+ L+Q AWN ND LRT+
Sbjct: 465 VDLKTELSRTERHILKEMGFICHVEHPHKFISNYLATLETPPE-LSQEAWNLANDSLRTT 641
Query: 209 LC 210
LC
Sbjct: 642 LC 647
>TC83138 similar to GP|9759604|dbj|BAB11392.1 contains similarity to
cyclin~gene_id:K18C1.7 {Arabidopsis thaliana}, partial
(6%)
Length = 1082
Score = 96.7 bits (239), Expect = 2e-20
Identities = 102/332 (30%), Positives = 159/332 (47%), Gaps = 36/332 (10%)
Frame = +3
Query: 323 SSANHVGPVS---NHGTNGSTEM-----IHLVEGDAKGNQHPEQEPPPHKENLQEAQDTV 374
S N+ PVS N +GS EM H ++ + + +Q E+ P K+ L Q+ +
Sbjct: 60 SVENNAVPVSRTENQSNDGSAEMGSDITDHKMDSETRDSQTSEKLPD--KDKLVGDQEKL 233
Query: 375 RSRSGYSEEPEINIGRSNMKEDVELNDK--HSSKNLNHRDGTFAPPSLEAIKKIDRDKVK 432
+ +E +GR+ +D LN + +NL R+G EAIK ID+DKVK
Sbjct: 234 VATKEAAE-----LGRN---DDTSLNKSSFNVGQNLERREGPLGQSPKEAIK-IDKDKVK 386
Query: 433 AALEKRKKAVGNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDED----LIERV 488
A LEKR+K G +T K ++MD++DL E RELEDG+E+ NK D+D +E++
Sbjct: 387 AILEKRRKERGEMTIKKDVMDEDDLIE--RELEDGVEIT-----NKMIDKDKVKTALEKM 545
Query: 489 RELEDGIEL----------------------AAQSEKNKQDGQKSLSKPVDRSAFENMQQ 526
R+ + + ++EKNKQ ++SLSKP D
Sbjct: 546 RKERGEMTIKKRYNGRG*SH*EGALKME*S*QLRNEKNKQQRRQSLSKPDD--------- 698
Query: 527 GKHQDHSEEHLHLVKPSHETDLSAVEEGEVSALDDIDLGRKSSNHKRKAESSPDKFAEGK 586
+DH E+ ++ + + D E+G++ +DD +++HKRK S P E K
Sbjct: 699 ---EDHGED----LEEARDRDGFNAEKGDM--IDDAS-SLLNNHHKRKG-SPPASQPETK 845
Query: 587 KRHNYSFGSTHHNRIDYIEDQNNVNLLGHAER 618
KR S +HN D+ E N + G+A+R
Sbjct: 846 KR----LDSNYHN--DFSE*GNGKDRFGYADR 923
>BG648100
Length = 526
Score = 73.6 bits (179), Expect = 2e-13
Identities = 36/85 (42%), Positives = 54/85 (63%)
Frame = +2
Query: 101 IIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKEVYEQQKELILLGER 160
+IAT+ +FLAGK EE+P PL V+ S E++HK+D A + + +EQ +E +L E+
Sbjct: 230 LIATAALFLAGKSEESPCPLNSVLRTSSELLHKQDFAFLSYWFPVDWFEQYRERVLEAEQ 409
Query: 161 VVLATLGFDFHVQHPYKPLVEAIKK 185
++L TL F+ VQH Y PL + K
Sbjct: 410 LILTTLNFELGVQHSYAPLTSVLNK 484
Score = 42.7 bits (99), Expect = 4e-04
Identities = 16/28 (57%), Positives = 24/28 (85%)
Frame = +1
Query: 72 KVPQVTIASAIIFCHRFFLRQSHAKNDR 99
K+PQ TI ++++ CHRFF+R+SHA +DR
Sbjct: 1 KMPQTTIGTSMVLCHRFFVRRSHACHDR 84
>CA858397 similar to GP|11340587|em unnamed protein product {Zea mays},
partial (33%)
Length = 870
Score = 68.9 bits (167), Expect = 5e-12
Identities = 37/98 (37%), Positives = 55/98 (55%), Gaps = 2/98 (2%)
Frame = +3
Query: 167 GFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVNDGLRTSLCLQFKPQHIAAGAIFLA 226
GF HV+HP+K + + L+ L Q AWN ND LRTSLC++FK + +A G ++ A
Sbjct: 3 GFICHVEHPHKFISNYLATLETPPE-LRQEAWNLANDSLRTSLCVRFKSEIVACGVVYAA 179
Query: 227 AKFLNVKLEK--LWWQVFDVTLTAHQLEEVSNQMLELY 262
A+ V L + WW+ FD + ++EV + LY
Sbjct: 180 ARRFQVPLPENPPWWKAFDAEKSG--IDEVCRVLAHLY 287
>BI262918 similar to GP|13365979|dbj putative cyclin homolog {Oryza sativa
(japonica cultivar-group)}, partial (21%)
Length = 377
Score = 67.0 bits (162), Expect = 2e-11
Identities = 32/86 (37%), Positives = 58/86 (67%)
Frame = +3
Query: 30 WYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFF 89
+YL+ +++ NSPS++DG+D E LR C +Q+ G+ L++PQ +A+ + HRF+
Sbjct: 123 FYLTDEQLT-NSPSRKDGIDEATETSLRIYGCDLIQESGILLRLPQAVMATGQVLFHRFY 299
Query: 90 LRQSHAKNDRRIIATSCMFLAGKVEE 115
++S A+ + + +A SC++LA K+EE
Sbjct: 300 CKKSFARFNVKKVAASCVWLASKLEE 377
>BF644639 similar to GP|9759604|dbj| contains similarity to
cyclin~gene_id:K18C1.7 {Arabidopsis thaliana}, partial
(4%)
Length = 700
Score = 62.0 bits (149), Expect = 7e-10
Identities = 27/33 (81%), Positives = 31/33 (93%)
Frame = -1
Query: 75 QVTIASAIIFCHRFFLRQSHAKNDRRIIATSCM 107
QV+IA+AIIFCHRFFLRQSHAKNDRR+I SC+
Sbjct: 436 QVSIATAIIFCHRFFLRQSHAKNDRRVIVISCL 338
>CA921228
Length = 822
Score = 57.0 bits (136), Expect = 2e-08
Identities = 33/60 (55%), Positives = 42/60 (70%), Gaps = 2/60 (3%)
Frame = -2
Query: 205 LRTSLCLQFKPQHIAAGAIFLAAKFLNVKLE--KLWWQVFDVTLTAHQLEEVSNQMLELY 262
LR+SL LQFKP IAAGA +LAAKFLN+ L K WQ F T + L++VS Q++EL+
Sbjct: 383 LRSSLWLQFKPHQIAAGAAYLAAKFLNMDLATYKNIWQEFQATPSV--LQDVSQQLMELF 210
>AW691348 weakly similar to GP|12744989|gb| putative cyclin {Arabidopsis
thaliana}, partial (9%)
Length = 618
Score = 42.4 bits (98), Expect = 5e-04
Identities = 16/31 (51%), Positives = 25/31 (80%)
Frame = +2
Query: 31 YLSRKEIEENSPSKQDGVDLKKEAYLRKSYC 61
++SR +I+ NSPS++DG+D+ E +LR SYC
Sbjct: 524 FMSRDDIDRNSPSRKDGIDVLHETHLRYSYC 616
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 36.6 bits (83), Expect = 0.030
Identities = 70/324 (21%), Positives = 124/324 (37%), Gaps = 12/324 (3%)
Frame = +3
Query: 268 APSNDVEGTAGGGNRATPKASTSNDEAATTNNNLYIRGPSPRLEASKLAISKNVVSSANH 327
APS G +A P+ ++ ++ +++++ E K +SK V S N
Sbjct: 348 APSKKTPAKKGNVKKAQPETTSEESDSDSSSSDE---------EEVKKPVSK-AVPSKNG 497
Query: 328 VGPVSNHGTNGSTEMIHLVEGDAKGNQHPEQEPPPHKENLQEAQDTVRSRSGYSEEPEIN 387
P T+ + + D K P + P K A+ E +
Sbjct: 498 SAPAKKVDTSEEEDSEESSDEDKK----PAAKAVPSKNGSAPAKKAASDEEDTDESSD-- 659
Query: 388 IGRSNMKEDVELNDKHSSKNLNHRDGTFAPPSLEAIKKIDRDKVKAALEKRK---KAVGN 444
ED E ++K ++K + ++G+ P + D D + E +K KA N
Sbjct: 660 -------EDEE-DEKPAAKAVPSKNGS-VPAKKADTESSDEDSESSDEEDKKPAAKASKN 812
Query: 445 I---TKKTELMDDEDLFERVRELEDGIEL---AAQSEKNKQDDEDLIERVRELEDGIELA 498
+ TKK DE+ E E ED + AA ++K+K+D D D +
Sbjct: 813 VSAPTKKAASSSDEESDEESDEDEDAKPVSKPAAVAKKSKKDSSD--------SDDEDDD 968
Query: 499 AQSEKNKQDGQKSLSKPVDRSAFENM---QQGKHQDHSEEHLHLVKPSHETDLSAVEEGE 555
+ S+++K KPV + M + G D SEE E + S + +
Sbjct: 969 SSSDEDK--------KPVAAKKEDKMNVDKDGSDSDQSEE-------ESEDEPSKTPQKK 1103
Query: 556 VSALDDIDLGRKSSNHKRKAESSP 579
+ ++ +D G+ +KA ++P
Sbjct: 1104IKDVEMVDAGKSG----KKAPNTP 1163
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 36.6 bits (83), Expect = 0.030
Identities = 36/151 (23%), Positives = 66/151 (42%), Gaps = 1/151 (0%)
Frame = +1
Query: 432 KAALEKRK-KAVGNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDEDLIERVRE 490
K+ LE + K G +++ +++E+ E E+ L ++E+NK D+E + E
Sbjct: 7 KSHLENEESKETGESSEEKSHLENEESKETGESSEEKSHL--ENEENK-DEEKSKQENEE 177
Query: 491 LEDGIELAAQSEKNKQDGQKSLSKPVDRSAFENMQQGKHQDHSEEHLHLVKPSHETDLSA 550
++DG ++ ++E+NK + E QQ ++ EE K E +L
Sbjct: 178 IKDGEKIQQENEENKDE--------------EKSQQENEENKDEE-----KSQQENELKK 300
Query: 551 VEEGEVSALDDIDLGRKSSNHKRKAESSPDK 581
E GE + + K N + +S DK
Sbjct: 301 NEGGEKETGEITEEKSKQENEETSETNSKDK 393
Score = 33.1 bits (74), Expect = 0.33
Identities = 31/152 (20%), Positives = 65/152 (42%), Gaps = 11/152 (7%)
Frame = +1
Query: 450 ELMDDEDLFERVRELEDGIELAAQSEKNK-------QDDEDLIERVRELEDGIELAAQSE 502
E D+E + E++DG ++ ++E+NK +++E+ E + E+ ++ E
Sbjct: 136 ENKDEEKSKQENEEIKDGEKIQQENEENKDEEKSQQENEENKDEEKSQQENELKKNEGGE 315
Query: 503 KNKQDGQKSLSKPVDRSAFENMQQGKHQDHSEEH----LHLVKPSHETDLSAVEEGEVSA 558
K + + SK + E + K + S ++ V +HE D E
Sbjct: 316 KETGEITEEKSKQENEETSETNSKDKENEESNQNGSDAKEQVGENHEQDSKQGTE----E 483
Query: 559 LDDIDLGRKSSNHKRKAESSPDKFAEGKKRHN 590
+ + G K + K K ++S D + +++N
Sbjct: 484 TNGTEGGEKEEHDKIKEDTSSDNQVQDGEKNN 579
>TC89772 similar to GP|9759529|dbj|BAB10995.1 gene_id:K21L19.3~unknown
protein {Arabidopsis thaliana}, partial (6%)
Length = 1230
Score = 35.0 bits (79), Expect = 0.087
Identities = 48/213 (22%), Positives = 88/213 (40%)
Frame = +1
Query: 311 EASKLAISKNVVSSANHVGPVSNHGTNGSTEMIHLVEGDAKGNQHPEQEPPPHKENLQEA 370
+ + L +SK + + N GP + G S GD + +H E P H+ + ++
Sbjct: 13 DLNSLGLSKKSLENPN--GPSNEKGLKES--------GDQERAEH---EIPGHRLS-RKH 150
Query: 371 QDTVRSRSGYSEEPEINIGRSNMKEDVELNDKHSSKNLNHRDGTFAPPSLEAIKKIDRDK 430
QD + S E+ + GRS ++ ++ S N P S K ID++
Sbjct: 151 QDGISSDD---EQQDSYRGRSKLERWTSHKERDFSIN--------KPSSSLKFKDIDKNN 297
Query: 431 VKAALEKRKKAVGNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDEDLIERVRE 490
+ E K V K +L + + L R+ D S+++ D +ER+++
Sbjct: 298 NGGSSEAGKP-VDESAKTVDLDNQQPLMPEARDSVDTESRDGDSKESGDRHLDTVERLKK 474
Query: 491 LEDGIELAAQSEKNKQDGQKSLSKPVDRSAFEN 523
+ +L SEK +K ++P+ + EN
Sbjct: 475 RSERFKLPMPSEKETLVIKKLETEPLPSAKTEN 573
>TC85960 similar to GP|23094138|gb|AAF50778.2 CG4835-PA {Drosophila
melanogaster}, partial (2%)
Length = 1834
Score = 35.0 bits (79), Expect = 0.087
Identities = 38/159 (23%), Positives = 61/159 (37%), Gaps = 19/159 (11%)
Frame = +3
Query: 289 TSNDEAATTNNNLYIRGPSPRLEASKLAISKNVVSSANHVGPVSNHGTN----------- 337
T D + NNNL++ P R + ++ + H+ P H +
Sbjct: 1176 TITDLPESPNNNLHLPLPETRPHTAPPSLHNQHGETLQHITPPPPHKKHKPPSQNPTAPV 1355
Query: 338 GSTEMIHLVEGDAKGNQHPEQEPPP---HKENLQEAQDTVRSRSGYSEEPEINIGRSNMK 394
+E+ H A+ P Q+ PP K N Q+ D R + G S E G ++
Sbjct: 1356 AKSEVAHAA---AECGLLPTQQQPP*GHTKTNQQQQLDDRRKQPG-SRRNETTTGPPQIR 1523
Query: 395 EDVEL-----NDKHSSKNLNHRDGTFAPPSLEAIKKIDR 428
N H + +H+ T PP+ +AI K+DR
Sbjct: 1524 TATTRPPQIRNIHHHLTSPDHQPLTTGPPTDQAINKLDR 1640
>TC81784 similar to PIR|T40294|T40294 hypothetical protein SPBC354.14c -
fission yeast (Schizosaccharomyces pombe), partial (30%)
Length = 1029
Score = 31.2 bits (69), Expect = 1.3
Identities = 18/35 (51%), Positives = 23/35 (65%), Gaps = 2/35 (5%)
Frame = +1
Query: 470 LAAQSEKNKQD--DEDLIERVRELEDGIELAAQSE 502
LAA SE+NK+ D +ERVREL + L+ QSE
Sbjct: 271 LAASSERNKRAIVDAGAVERVRELVLNVPLSVQSE 375
>TC76817 GP|2598577|emb|CAA75576.1 MtN22 {Medicago truncatula}, complete
Length = 795
Score = 30.8 bits (68), Expect = 1.6
Identities = 28/105 (26%), Positives = 51/105 (47%), Gaps = 4/105 (3%)
Frame = +3
Query: 451 LMDDEDLFERVRELEDGIELAAQSEK-NKQDDEDLIERVRELEDGIELAAQSEK-NKQDG 508
L D+ + F + E ED + L+ + K +K +ED + + E ED + + ++ +K DG
Sbjct: 351 LCDEGNKFSEIDE-EDNVSLSDREYKASKSGEEDGVSKSNE-EDKVSKNDEGDQVSKIDG 524
Query: 509 QKSLSK--PVDRSAFENMQQGKHQDHSEEHLHLVKPSHETDLSAV 551
+ ++SK D NM+ + E+ + L K S T+ V
Sbjct: 525 EDAVSKNDKEDNVFMSNMEDNVFKSDEEDKISLTK*SS*TNYQKV 659
>TC76815 MtN22
Length = 795
Score = 30.8 bits (68), Expect = 1.6
Identities = 28/105 (26%), Positives = 51/105 (47%), Gaps = 4/105 (3%)
Frame = +1
Query: 451 LMDDEDLFERVRELEDGIELAAQSEK-NKQDDEDLIERVRELEDGIELAAQSEK-NKQDG 508
L D+ + F + E ED + L+ + K +K +ED + + E ED + + ++ +K DG
Sbjct: 328 LCDEGNKFSEIDE-EDNVSLSDREYKASKSGEEDGVSKSNE-EDKVSKNDEGDQVSKIDG 501
Query: 509 QKSLSK--PVDRSAFENMQQGKHQDHSEEHLHLVKPSHETDLSAV 551
+ ++SK D NM+ + E+ + L K S T+ V
Sbjct: 502 EDAVSKNDKEDNVFMSNMEDNVFKSDEEDKISLTK*SS*TNYQKV 636
>TC76816 GP|2598577|emb|CAA75576.1 MtN22 {Medicago truncatula}, partial
(83%)
Length = 661
Score = 30.8 bits (68), Expect = 1.6
Identities = 28/105 (26%), Positives = 51/105 (47%), Gaps = 4/105 (3%)
Frame = +2
Query: 451 LMDDEDLFERVRELEDGIELAAQSEK-NKQDDEDLIERVRELEDGIELAAQSEK-NKQDG 508
L D+ + F + E ED + L+ + K +K +ED + + E ED + + ++ +K DG
Sbjct: 218 LCDEGNKFSEIDE-EDNVSLSDREYKASKSGEEDGVSKSNE-EDKVSKNDEGDQVSKIDG 391
Query: 509 QKSLSK--PVDRSAFENMQQGKHQDHSEEHLHLVKPSHETDLSAV 551
+ ++SK D NM+ + E+ + L K S T+ V
Sbjct: 392 EDAVSKNDKEDNVFMSNMEDNVFKSDEEDKISLTK*SS*TNYQKV 526
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.311 0.130 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,519,248
Number of Sequences: 36976
Number of extensions: 229167
Number of successful extensions: 1089
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 1045
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1075
length of query: 629
length of database: 9,014,727
effective HSP length: 102
effective length of query: 527
effective length of database: 5,243,175
effective search space: 2763153225
effective search space used: 2763153225
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0176a.15


