

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
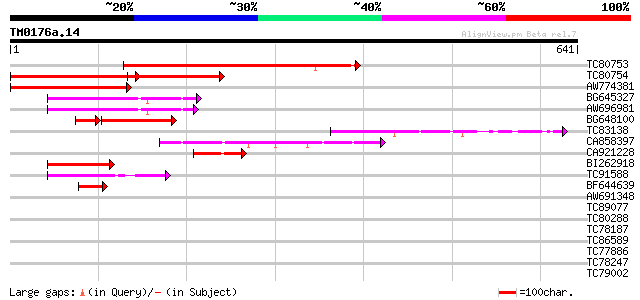
Query= TM0176a.14
(641 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80753 homologue to PIR|T06153|T06153 hypothetical protein F24J... 412 e-115
TC80754 homologue to GP|9759604|dbj|BAB11392.1 contains similari... 242 3e-64
AW774381 homologue to GP|9759604|dbj contains similarity to cycl... 225 4e-59
BG645327 similar to GP|13924511|gb ania-6a type cyclin {Arabidop... 107 1e-23
AW696981 similar to GP|13924511|gb ania-6a type cyclin {Arabidop... 103 2e-22
BG648100 75 8e-14
TC83138 similar to GP|9759604|dbj|BAB11392.1 contains similarity... 72 9e-13
CA858397 similar to GP|11340587|em unnamed protein product {Zea ... 71 1e-12
CA921228 67 2e-11
BI262918 similar to GP|13365979|dbj putative cyclin homolog {Ory... 65 6e-11
TC91588 similar to SP|P93411|CG1C_ORYSA G1/S-specific cyclin C-t... 61 1e-09
BF644639 similar to GP|9759604|dbj| contains similarity to cycli... 61 1e-09
AW691348 weakly similar to GP|12744989|gb| putative cyclin {Arab... 42 0.001
TC89077 weakly similar to GP|21740511|emb|CAD41490. OSJNBa0029H0... 38 0.014
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 36 0.040
TC78187 homologue to GP|7381225|gb|AAF61443.1| root border cell-... 35 0.068
TC86589 similar to GP|7576200|emb|CAB87861.1 protein synthesis i... 35 0.12
TC77886 similar to GP|14532516|gb|AAK63986.1 AT5g05210/K2A11_8 {... 34 0.15
TC78247 similar to GP|15451184|gb|AAK96863.1 putative protein {A... 33 0.26
TC79002 similar to GP|15375305|gb|AAK54745.2 putative transcript... 32 0.58
>TC80753 homologue to PIR|T06153|T06153 hypothetical protein F24J7.161 -
Arabidopsis thaliana, partial (27%)
Length = 900
Score = 412 bits (1060), Expect = e-115
Identities = 217/274 (79%), Positives = 230/274 (83%), Gaps = 6/274 (2%)
Frame = +1
Query: 129 VSYEIIHKKDPAAVQRIKQKDVYEQQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKK 188
VSYEII+KKDP AVQRIKQK+VYEQQKELILL ERVVLATLGFD NVHHPYKPLVEAIKK
Sbjct: 1 VSYEIINKKDPTAVQRIKQKEVYEQQKELILLAERVVLATLGFDFNVHHPYKPLVEAIKK 180
Query: 189 FKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQE 248
FKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQE
Sbjct: 181 FKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQE 360
Query: 249 FDVTPRQLEEVSNQMLELYEQNRMAQSNDVEGTTGGGN--RATPKAPTSNDEAASTNRNL 306
FDVTPRQLEEVSNQMLELYEQNRM SN+ EGTTGG R T KAPTSNDE A N N
Sbjct: 361 FDVTPRQLEEVSNQMLELYEQNRMPPSNEAEGTTGGAASIRTTVKAPTSNDETAPANSNS 540
Query: 307 HIGGPSSTLETSKPATSKNVFVSS-ANHVGRPVSNHGVS---GSTEVKHPVEDDVKGNLH 362
GG SS LETSKPA+SK F SS AN VGRPVSN G S GSTE+KH VEDDVKGN H
Sbjct: 541 QTGGISSRLETSKPASSKTAFDSSAANQVGRPVSNPGRSRDYGSTEIKHRVEDDVKGNQH 720
Query: 363 PKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQ 396
P+ EP Y++N+Q+ Q++VRSRS G + KQ
Sbjct: 721 PEHEPLPYKDNLQQVQNVVRSRSD--NGQKSTKQ 816
>TC80754 homologue to GP|9759604|dbj|BAB11392.1 contains similarity to
cyclin~gene_id:K18C1.7 {Arabidopsis thaliana}, partial
(38%)
Length = 1115
Score = 242 bits (618), Expect = 3e-64
Identities = 122/149 (81%), Positives = 132/149 (87%), Gaps = 1/149 (0%)
Frame = +2
Query: 1 MAGLLLGDISHHGSYQSGSQGGSQDNQEYGSRWYFSRKEIEENSPSKQDGVDLKKEAYLR 60
MAGLLLGD SH G++ GSQ S++ E GSRWYFSRKEIEENSPS++DG+DLKKE YLR
Sbjct: 203 MAGLLLGDASHQGAHPVGSQRYSEEKPEDGSRWYFSRKEIEENSPSQEDGIDLKKETYLR 382
Query: 61 KSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEETP 120
KSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRR IATVCMFLAGKVEETP
Sbjct: 383 KSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRTIATVCMFLAGKVEETP 562
Query: 121 RPLKDVILVSYEIIHK-KDPAAVQRIKQK 148
RPLKDVI+VSYEII+K + QRIKQK
Sbjct: 563 RPLKDVIMVSYEIINKERIQQQFQRIKQK 649
Score = 191 bits (484), Expect = 1e-48
Identities = 95/110 (86%), Positives = 100/110 (90%)
Frame = +3
Query: 134 IHKKDPAAVQRIKQKDVYEQQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQ 193
I K ++ + +K+VYEQQKELILL ERVVLATLGFD NVHHPYKPLVEAIKKFKVAQ
Sbjct: 606 IKKGSNSSFRESNRKEVYEQQKELILLAERVVLATLGFDFNVHHPYKPLVEAIKKFKVAQ 785
Query: 194 NALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEK 243
NALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEK
Sbjct: 786 NALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEK 935
Score = 37.7 bits (86), Expect = 0.014
Identities = 16/24 (66%), Positives = 18/24 (74%)
Frame = +2
Query: 234 KVKLPSDGEKVWWQEFDVTPRQLE 257
K + P +VWWQEFDVTPRQLE
Sbjct: 905 KGEAPIGWGEVWWQEFDVTPRQLE 976
>AW774381 homologue to GP|9759604|dbj contains similarity to
cyclin~gene_id:K18C1.7 {Arabidopsis thaliana}, partial
(18%)
Length = 628
Score = 225 bits (574), Expect = 4e-59
Identities = 110/138 (79%), Positives = 120/138 (86%), Gaps = 1/138 (0%)
Frame = +1
Query: 1 MAGLLLGDISHHGSYQSGSQGGSQDNQEYG-SRWYFSRKEIEENSPSKQDGVDLKKEAYL 59
MA L+ G++ HHG+ S SQD QE RWY SRKEIEEN+PS++DG+DLKKE YL
Sbjct: 214 MAALVSGELPHHGASDGSSCRSSQDKQEEALGRWYMSRKEIEENAPSRKDGIDLKKETYL 393
Query: 60 RKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEET 119
RKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRR IATVCMFLAGKVEET
Sbjct: 394 RKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRTIATVCMFLAGKVEET 573
Query: 120 PRPLKDVILVSYEIIHKK 137
PRPLKDVIL+SYE+IHKK
Sbjct: 574 PRPLKDVILISYEMIHKK 627
>BG645327 similar to GP|13924511|gb ania-6a type cyclin {Arabidopsis
thaliana}, partial (51%)
Length = 789
Score = 107 bits (268), Expect = 1e-23
Identities = 57/179 (31%), Positives = 104/179 (57%), Gaps = 4/179 (2%)
Frame = +1
Query: 43 NSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDR 102
NSPS++DG+D E LR C +Q+ G+ L++PQ +AT + HRF+ ++S A+ +
Sbjct: 199 NSPSRKDGIDEATETTLRIYGCDLIQESGILLRLPQAVMATGQVLFHRFYCKKSFARFNV 378
Query: 103 RIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYEQQ----KELI 158
+ +A C++LA K+EE R + V+++ + + +++ + + D+Y ++ K +
Sbjct: 379 KKVAASCVWLASKLEENTRKARQVLIIFHRMECRRENLPIDHL---DLYSKKYVDLKTEL 549
Query: 159 LLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFK 217
ER +L +GF +V HP+K + + + L+Q AWN ND LRT+LC++F+
Sbjct: 550 SRTERHILKEMGFICHVEHPHKFISNYLATLETPPE-LSQEAWNLANDSLRTTLCVRFR 723
>AW696981 similar to GP|13924511|gb ania-6a type cyclin {Arabidopsis
thaliana}, partial (45%)
Length = 648
Score = 103 bits (258), Expect = 2e-22
Identities = 56/175 (32%), Positives = 100/175 (57%), Gaps = 4/175 (2%)
Frame = +3
Query: 43 NSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDR 102
NSPS++DG+D E LR C +Q+ G+ L++PQ +AT + HRF+ ++S A+ +
Sbjct: 135 NSPSRKDGIDEATETTLRIYGCDLIQESGILLRLPQAVMATGQVLFHRFYCKKSFARFNV 314
Query: 103 RIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYEQQ----KELI 158
+ +A C++LA K+EE R + V+++ + + +++ + + D+Y ++ K +
Sbjct: 315 KKVAASCVWLASKLEENTRKARQVLIIFHRMECRRENLPIDHL---DLYSKKYVDLKTEL 485
Query: 159 LLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLC 213
ER +L +GF +V HP+K + + + L+Q AWN ND LRT+LC
Sbjct: 486 SRTERHILKEMGFICHVEHPHKFISNYLATLETPPE-LSQEAWNLANDSLRTTLC 647
>BG648100
Length = 526
Score = 75.1 bits (183), Expect = 8e-14
Identities = 37/85 (43%), Positives = 53/85 (61%)
Frame = +2
Query: 104 IIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYEQQKELILLGER 163
+IAT +FLAGK EE+P PL V+ S E++HK+D A + D +EQ +E +L E+
Sbjct: 230 LIATAALFLAGKSEESPCPLNSVLRTSSELLHKQDFAFLSYWFPVDWFEQYRERVLEAEQ 409
Query: 164 VVLATLGFDLNVHHPYKPLVEAIKK 188
++L TL F+L V H Y PL + K
Sbjct: 410 LILTTLNFELGVQHSYAPLTSVLNK 484
Score = 44.3 bits (103), Expect = 1e-04
Identities = 17/28 (60%), Positives = 24/28 (85%)
Frame = +1
Query: 75 KVPQVTIATAIIFCHRFFLRQSHAKNDR 102
K+PQ TI T+++ CHRFF+R+SHA +DR
Sbjct: 1 KMPQTTIGTSMVLCHRFFVRRSHACHDR 84
>TC83138 similar to GP|9759604|dbj|BAB11392.1 contains similarity to
cyclin~gene_id:K18C1.7 {Arabidopsis thaliana}, partial
(6%)
Length = 1082
Score = 71.6 bits (174), Expect = 9e-13
Identities = 82/322 (25%), Positives = 131/322 (40%), Gaps = 54/322 (16%)
Frame = +3
Query: 363 PKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQ 422
P Q S + ++ +S G A+ E + + + + + +Q
Sbjct: 45 PPQRSSVENNAVPVSRTENQSNDGSAEMGSDITDHKMDSETRDSQTSEKLPDKDKLVGDQ 224
Query: 423 ESNVGRSNIKE----EDVELNDK--HSSKNLNHRDATFASPNLEGIKKIDRDKVKAALEK 476
E V E +D LN + +NL R+ E IK ID+DKVKA LEK
Sbjct: 225 EKLVATKEAAELGRNDDTSLNKSSFNVGQNLERREGPLGQSPKEAIK-IDKDKVKAILEK 401
Query: 477 RKKATGNISKKTELVDDEDLIERVRELEDGIELA-------------------------- 510
R+K G ++ K +++D++DLIE RELEDG+E+
Sbjct: 402 RRKERGEMTIKKDVMDEDDLIE--RELEDGVEITNKMIDKDKVKTALEKMRKERGEMTIK 575
Query: 511 ----------------------AQSEKNKQDGRKSLSKSLDRSAFENMQHGKHQDHSEEH 548
++EKNKQ R+SLSK D +DH E+
Sbjct: 576 KRYNGRG*SH*EGALKME*S*QLRNEKNKQQRRQSLSKPDD------------EDHGED- 716
Query: 549 LHLVKPSHEADLSAVEEGEVSALDDIDLGPKSSNQKRKVESSPDNFVEGKKRHNYGSGST 608
++ + + D E+G++ +D D +N ++ S P + E KKR + S
Sbjct: 717 ---LEEARDRDGFNAEKGDM--ID--DASSLLNNHHKRKGSPPASQPETKKRLD----SN 863
Query: 609 HHNRIDYIEDRNKVNRLGHAER 630
+HN D+ E N +R G+A+R
Sbjct: 864 YHN--DFSE*GNGKDRFGYADR 923
>CA858397 similar to GP|11340587|em unnamed protein product {Zea mays},
partial (33%)
Length = 870
Score = 71.2 bits (173), Expect = 1e-12
Identities = 65/284 (22%), Positives = 117/284 (40%), Gaps = 28/284 (9%)
Frame = +3
Query: 170 GFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLA 229
GF +V HP+K + + + L Q AWN ND LRTSLC++FK +A G ++ A
Sbjct: 3 GFICHVEHPHKFISNYLATLETPPE-LRQEAWNLANDSLRTSLCVRFKSEIVACGVVYAA 179
Query: 230 AKFLKVKLPSDGEKVWWQEFDVTPRQLEEVSNQMLELYE------------------QNR 271
A+ +V LP + WW+ FD ++EV + LY N+
Sbjct: 180 ARRFQVPLPEN--PPWWKAFDAEKSGIDEVCRVLAHLYSLPKAQYIPVCKDEDSFTFSNK 353
Query: 272 MAQSN--DVEGTTGGGNRATPKAPTSNDEA---ASTNRNLHIGGPSSTLETSKPATSKNV 326
+S DV ++ N TP + +EA S+++ + S L ++ + ++
Sbjct: 354 SLESKAMDVPQSSSPTNAETPALKGALEEANIDLSSSKGALVKQTSDKLNDARKSDDESK 533
Query: 327 FVSSANHVG-----RPVSNHGVSGSTEVKHPVEDDVKGNLHPKQEPSAYEENMQEAQDMV 381
++ V + S+ V S +++ + D +K + S E +EA+
Sbjct: 534 GTAAERDVKDELTFKSKSDRRVEASGDLRRDL-DRIKSRGRGRGRDSDNEREREEAERFK 710
Query: 382 RSRSGYAKGNQHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESN 425
G+ Q + + + K + D RS Y ++ +
Sbjct: 711 LKDHGH-HSRQRARDSGHSEKSKRHSSHDRDYYRSLYSVREKGS 839
>CA921228
Length = 822
Score = 67.4 bits (163), Expect = 2e-11
Identities = 35/60 (58%), Positives = 44/60 (73%)
Frame = -2
Query: 208 LRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQEFDVTPRQLEEVSNQMLELY 267
LR+SL LQFKPH IAAGA +LAAKFL + L + K WQEF TP L++VS Q++EL+
Sbjct: 383 LRSSLWLQFKPHQIAAGAAYLAAKFLNMDLAT--YKNIWQEFQATPSVLQDVSQQLMELF 210
>BI262918 similar to GP|13365979|dbj putative cyclin homolog {Oryza sativa
(japonica cultivar-group)}, partial (21%)
Length = 377
Score = 65.5 bits (158), Expect = 6e-11
Identities = 30/76 (39%), Positives = 50/76 (65%)
Frame = +3
Query: 43 NSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDR 102
NSPS++DG+D E LR C +Q+ G+ L++PQ +AT + HRF+ ++S A+ +
Sbjct: 150 NSPSRKDGIDEATETSLRIYGCDLIQESGILLRLPQAVMATGQVLFHRFYCKKSFARFNV 329
Query: 103 RIIATVCMFLAGKVEE 118
+ +A C++LA K+EE
Sbjct: 330 KKVAASCVWLASKLEE 377
>TC91588 similar to SP|P93411|CG1C_ORYSA G1/S-specific cyclin C-type. [Rice]
{Oryza sativa}, partial (63%)
Length = 658
Score = 61.2 bits (147), Expect = 1e-09
Identities = 40/140 (28%), Positives = 69/140 (48%)
Frame = +1
Query: 43 NSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDR 102
N K+ GV L+ ++ ++ L ++KV Q +ATA+ + R + R S + D
Sbjct: 214 NPVDKEKGVTLEDFKLIKMHMSNYILKLAQQVKVRQRVVATAVTYMRRVYTRMSMTEYDP 393
Query: 103 RIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYEQQKELILLGE 162
R++A C++LA K EE+ ++ +LV Y ++++ D Y + + IL E
Sbjct: 394 RLVAPACLYLASKAEES--TVQARLLVFY----------IKKLYADDKYRYEIKDILEME 537
Query: 163 RVVLATLGFDLNVHHPYKPL 182
+L L L V HPY+ L
Sbjct: 538 MKILEALKXYLVVFHPYRSL 597
>BF644639 similar to GP|9759604|dbj| contains similarity to
cyclin~gene_id:K18C1.7 {Arabidopsis thaliana}, partial
(4%)
Length = 700
Score = 61.2 bits (147), Expect = 1e-09
Identities = 27/33 (81%), Positives = 30/33 (90%)
Frame = -1
Query: 78 QVTIATAIIFCHRFFLRQSHAKNDRRIIATVCM 110
QV+IATAIIFCHRFFLRQSHAKNDRR+I C+
Sbjct: 436 QVSIATAIIFCHRFFLRQSHAKNDRRVIVISCL 338
>AW691348 weakly similar to GP|12744989|gb| putative cyclin {Arabidopsis
thaliana}, partial (9%)
Length = 618
Score = 41.6 bits (96), Expect = 0.001
Identities = 16/31 (51%), Positives = 24/31 (76%)
Frame = +2
Query: 34 YFSRKEIEENSPSKQDGVDLKKEAYLRKSYC 64
+ SR +I+ NSPS++DG+D+ E +LR SYC
Sbjct: 524 FMSRDDIDRNSPSRKDGIDVLHETHLRYSYC 616
>TC89077 weakly similar to GP|21740511|emb|CAD41490. OSJNBa0029H02.6 {Oryza
sativa}, partial (5%)
Length = 1472
Score = 37.7 bits (86), Expect = 0.014
Identities = 55/255 (21%), Positives = 102/255 (39%), Gaps = 40/255 (15%)
Frame = +1
Query: 284 GGNRATPKAPTSNDEA---ASTNRNLHIGGPSSTLETSKPATSKNVFVSSANHVGRPVS- 339
GGN++ K +S + ST + G S++ +T+K K A R S
Sbjct: 694 GGNKSNKKLKSSKSPSLHEVSTGKKQRSSGGSTSKKTTKIVNGKPSPSKHAKRGQRKASK 873
Query: 340 ---NHGV--SGSTEVKHPVE----------DDVKG--------NLHPKQEPSAYEENMQE 376
+ GV S +E+ +P E D KG N+ K++P+ +++
Sbjct: 874 TNFHEGVNESSDSELPNPEETTIAEAEINSDGSKGEQDEGSDVNITKKKKPNRKRKSVSW 1053
Query: 377 AQDMVRSRSGYAKGN--------QHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESNVGR 428
+ + +S K KQEP EK + + R GY + +++
Sbjct: 1054GKRSKKKKSVSNKKEPDEEKHEPNEEKQEPDA--EKQDYPETLSEDREGYPQGAQNDEEE 1227
Query: 429 SNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKAALEKRKKATGNISKK- 487
++ KE D + + + S +N+N + + N + + D + + KK+ +++
Sbjct: 1228NSSKERDADESREASRENVNEEEESGPEGN-----QHESDVESSPSREVKKSLDDLTSPE 1392
Query: 488 ----TELVDDEDLIE 498
EL DDE LI+
Sbjct: 1393DATFAELPDDEPLIK 1437
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100
- Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 36.2 bits (82), Expect = 0.040
Identities = 49/228 (21%), Positives = 90/228 (38%), Gaps = 6/228 (2%)
Frame = +3
Query: 351 HP--VEDDVKGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQEPSTYE-EKMQE 407
HP VE D ++E NMQ ++ + +GN+H +E S + +E
Sbjct: 363 HPGKVEADKNEGHEEEEEDEHIVYNMQNKREHDEQQQEGEEGNKHETEEESEDNVHERRE 542
Query: 408 AQDMVRSRSGYGKEQESNVGRSNIKEE--DVELNDKHSSKNLNHRDATFASPNLEGIKKI 465
QD ++ G ++E+ +++E DVE+++ K+ +
Sbjct: 543 EQDEEENKHGAEVQEENESKSEEVEDEGGDVEIDENDHEKS-----------------EA 671
Query: 466 DRDKVKAALEKRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGRKSLS 525
D D+ +++ K +TE D ED E+ +E+ A+ E K D S
Sbjct: 672 DNDREDEVVDEEKDKEEEGDDETENEDKED-EEKGGLVENHENHEAREEHYKAD---DAS 839
Query: 526 KSLDRSAFENMQHGKHQDHSEEHL-HLVKPSHEADLSAVEEGEVSALD 572
++ E + +HS+ L + KP +E + S G + D
Sbjct: 840 SAVAHDTHETSTETGNLEHSDLSLQNTRKPENETNHSDESYGSQNVSD 983
Score = 33.5 bits (75), Expect = 0.26
Identities = 53/283 (18%), Positives = 94/283 (32%), Gaps = 58/283 (20%)
Frame = +3
Query: 350 KHPVEDDVKGNLHPKQEPSAYEENMQ--EAQDMVRSRSGYAKG---------NQHPKQEP 398
KH E++ + N+H ++E EEN E Q+ S+S + N H K E
Sbjct: 492 KHETEEESEDNVHERREEQDEEENKHGAEVQEENESKSEEVEDEGGDVEIDENDHEKSEA 671
Query: 399 STYEE---------KMQEAQDMVRSRSGYGKEQ--------------------------- 422
E K +E D + +E+
Sbjct: 672 DNDREDEVVDEEKDKEEEGDDETENEDKEDEEKGGLVENHENHEAREEHYKADDASSAVA 851
Query: 423 ----ESNVGRSNIKEEDVELNDKHSSKN-LNHRDATFASPNLEGIKKIDRDKVKAALEKR 477
E++ N++ D+ L + +N NH D ++ S N+ +K + +
Sbjct: 852 HDTHETSTETGNLEHSDLSLQNTRKPENETNHSDESYGSQNVSDLKVTEGELTDGVSSNA 1031
Query: 478 K--KATGN--ISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGRKSLSKSLDRSAF 533
K TGN S +T + ++ L I A+ + D S S+ +
Sbjct: 1032TAGKETGNDSFSNETAKTKPDSQLDLSSNLTAVITEASSNSSGTGDDTSSSSEQIKAVIL 1211
Query: 534 ENMQHGKHQDHSEEHLHLVKPSH--EADLSAVEEGEVSALDDI 574
H ++ + +K + E S + EG + D I
Sbjct: 1212SESDHAQNATVNTTITGDMKQTEGLEQSGSKISEGNLPGNDSI 1340
>TC78187 homologue to GP|7381225|gb|AAF61443.1| root border cell-specific
protein {Pisum sativum}, complete
Length = 1397
Score = 35.4 bits (80), Expect = 0.068
Identities = 31/131 (23%), Positives = 48/131 (35%), Gaps = 13/131 (9%)
Frame = -2
Query: 268 EQNRMAQSNDVEGTTGGGNRATPKAPTSNDEA-------ASTNRNLHIGGP---SSTLET 317
+ N+ + D TT G R K+P S A A+ +L G S
Sbjct: 685 DPNKAVATPDTNRTTFGSMRTNRKSPKSTQNALGCLAKYAARMASLSFSGTKIASPCKVI 506
Query: 318 SKPATSKNVFVSSANHVGRPVSNH---GVSGSTEVKHPVEDDVKGNLHPKQEPSAYEENM 374
+K S S H+G + S + + HP K + P PS + EN+
Sbjct: 505 TKSVLSSGSLAKSTEHLGLAAKSFE*TAKSLTANIGHPFASHAKSTIDPDG*PSYFFENV 326
Query: 375 QEAQDMVRSRS 385
+ +RSR+
Sbjct: 325 ESVPRRLRSRT 293
>TC86589 similar to GP|7576200|emb|CAB87861.1 protein synthesis initiation
factor-like {Arabidopsis thaliana}, partial (28%)
Length = 2539
Score = 34.7 bits (78), Expect = 0.12
Identities = 34/128 (26%), Positives = 61/128 (47%), Gaps = 1/128 (0%)
Frame = +3
Query: 462 IKKIDRDKVKAALEKRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGR 521
+K+ D ++ + + R++ GNI EL + L ER+ E +L Q + ++
Sbjct: 642 VKQSDEEREEKRTKARRRMLGNIRLIGELYKKKMLTERIMH-ECIKKLLGQCQNPDEEDI 818
Query: 522 KSLSKSLDRSAFENMQHGKHQDHSEEHLHLVK-PSHEADLSAVEEGEVSALDDIDLGPKS 580
++L K L + E + H K ++H + + +K S+ +LS+ D IDL
Sbjct: 819 EALCK-LMSTIGEMIDHPKAKEHMDVYFERIKLLSNNMNLSS--RLRFMLKDTIDLRKNR 989
Query: 581 SNQKRKVE 588
Q+RKVE
Sbjct: 990 WQQRRKVE 1013
>TC77886 similar to GP|14532516|gb|AAK63986.1 AT5g05210/K2A11_8 {Arabidopsis
thaliana}, partial (46%)
Length = 1439
Score = 34.3 bits (77), Expect = 0.15
Identities = 55/273 (20%), Positives = 104/273 (37%), Gaps = 7/273 (2%)
Frame = +3
Query: 234 KVKLPSDGEKVWWQEFDVTPRQLEEVSNQMLELYEQNRMAQSNDVEGTTGGGNRATPKAP 293
K LP+D EK W+Q + +Q E+ + ++ +++++ R P+ P
Sbjct: 276 KFYLPTDDEKPWFQ--GLNKKQKEKAKKKTIQNIKKSKV-------------QRLDPEKP 410
Query: 294 TSNDEAASTNRNLHIGGPSSTLETSKPATSKNVFVSSANHVGRPVSNHGVSGSTEVKHPV 353
+ E + G E + FVS + R V+
Sbjct: 411 VTTLELLKKSL-----GKEKANEGEEEEDVVKPFVSELDGDDRSVT-------------- 533
Query: 354 EDDVKGNLHPKQEPSAYEENMQEAQDMVRSRS------GYAKGNQHPKQEPSTYEEKMQE 407
++++ LH K E N + + + R GY + K++ +T E K
Sbjct: 534 YEELRQRLHRKLEGFRSTRNCADPEKAAKKREDRDTKRGYHYQDNKRKRDDATDESKPAP 713
Query: 408 AQDMVRSRSGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDR 467
+ + E + ++K +D E+ K K H++ A LE +KK D
Sbjct: 714 DEKLQEKVKKDAAEASKQLVFGHVKLQDEEMLGK-KRKISKHKELERAK-KLEELKKNDP 887
Query: 468 DKVKAALEKRK-KATGNISKKTELVDDEDLIER 499
+K +A +K KA + + ++ DD LI++
Sbjct: 888 EKGEAIAKKEAWKAAMDRASGIKVHDDPKLIKK 986
>TC78247 similar to GP|15451184|gb|AAK96863.1 putative protein {Arabidopsis
thaliana}, partial (15%)
Length = 1327
Score = 33.5 bits (75), Expect = 0.26
Identities = 41/182 (22%), Positives = 76/182 (41%), Gaps = 3/182 (1%)
Frame = +3
Query: 416 SGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKAALE 475
SG + +S+ N + ++ND ++S NLN R + N + I +I+ +K A+
Sbjct: 426 SGGSRRADSSKPEENFFSDLSKVNDDNASLNLNGRGFLVGNNNNKPI-EIEENKRSEAVN 602
Query: 476 KRKKA---TGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGRKSLSKSLDRSA 532
KR+ + N+ + + DL +R R A +DG + ++ + S
Sbjct: 603 KRRMSFDDIRNLKRHDSDIHHGDLHDRAR---------ASHISLTEDGSTAENEDVADSE 755
Query: 533 FENMQHGKHQDHSEEHLHLVKPSHEADLSAVEEGEVSALDDIDLGPKSSNQKRKVESSPD 592
EN +HS+ ++ +D S EV + D ++ QKR S+
Sbjct: 756 AENSTSRPLSNHSDGSKGFIRVGASSDASK----EVRGVAD----SSANGQKRFTGSTEK 911
Query: 593 NF 594
+F
Sbjct: 912 DF 917
>TC79002 similar to GP|15375305|gb|AAK54745.2 putative transcription factor
MYB124 {Arabidopsis thaliana}, partial (54%)
Length = 2304
Score = 32.3 bits (72), Expect = 0.58
Identities = 36/159 (22%), Positives = 69/159 (42%), Gaps = 10/159 (6%)
Frame = +3
Query: 416 SGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNHR-----DATFASPNLEGIKKIDRDKV 470
SGY Q + + + +K++D ++N L DA +LE KI ++ +
Sbjct: 1170 SGYA--QNNKIQGTFLKKDDPKINALMQQAELLTSLALKVDAENMDQSLENAWKILQEFM 1343
Query: 471 KAALEKRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGRKSL-----S 525
K +G +LVD +DL++ ++ + IE + + +D +S S
Sbjct: 1344 NR--NKESDVSGYKIPDLQLVDLKDLLQDLKNNSEEIEPCWRYMEFYEDSPESSEHSTGS 1517
Query: 526 KSLDRSAFENMQHGKHQDHSEEHLHLVKPSHEADLSAVE 564
+L EN++H HQD E L ++ H+ ++ +
Sbjct: 1518 TTLPHCTGENLEHSLHQDSGTE-LKSIQIEHQKEIGGCD 1631
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.311 0.129 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,301,048
Number of Sequences: 36976
Number of extensions: 216741
Number of successful extensions: 1079
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 1041
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1068
length of query: 641
length of database: 9,014,727
effective HSP length: 102
effective length of query: 539
effective length of database: 5,243,175
effective search space: 2826071325
effective search space used: 2826071325
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0176a.14


