

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0176a.13
(515 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
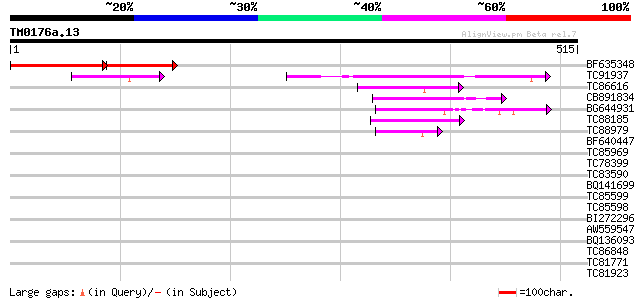
Score E
Sequences producing significant alignments: (bits) Value
BF635348 similar to GP|8439910|gb| It is a member of GTP1/OBG fa... 132 2e-48
TC91937 similar to GP|22136032|gb|AAM91598.1 GTP-binding protein... 142 3e-34
TC86616 similar to GP|7643796|gb|AAF65513.1| GTP-binding protein... 61 1e-09
CB891834 homologue to PIR|C96751|C96 probable GTP-binding protei... 59 6e-09
BG644931 similar to GP|22135878|gb GTP-binding protein putative... 54 2e-07
TC88185 homologue to PIR|T09368|T09368 GTP-binding protein F23K1... 53 3e-07
TC88979 similar to PIR|H96601|H96601 hypothetical protein T6H22.... 45 7e-05
BF640447 similar to GP|20152085|gb| RE21802p {Drosophila melanog... 36 0.031
TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsi... 32 0.59
TC78399 homologue to SP|O04834|SARA_ARATH GTP-binding protein SA... 31 1.0
TC83590 similar to GP|9758365|dbj|BAB08866.1 GTP binding protein... 31 1.3
BQ141699 similar to GP|334068|gb|A ORF2 {Pseudorabies virus}, pa... 31 1.3
TC85599 homologue to SP|O04834|SARA_ARATH GTP-binding protein SA... 30 1.7
TC85598 homologue to SP|O04834|SARA_ARATH GTP-binding protein SA... 30 2.9
BI272296 similar to GP|12322014|gb GTPase putative; 34281-30152... 29 3.8
AW559547 weakly similar to GP|12654451|gb| ADP-ribosylation fact... 29 5.0
BQ136093 29 5.0
TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {A... 28 6.5
TC81771 similar to GP|10176714|dbj|BAB09944. dimethylaniline mon... 28 6.5
TC81923 GP|21667854|gb|AAM74155.1 cyclin T1 protein {Capra hircu... 28 8.5
>BF635348 similar to GP|8439910|gb| It is a member of GTP1/OBG family
PF|01018. {Arabidopsis thaliana}, partial (5%)
Length = 545
Score = 132 bits (332), Expect(2) = 2e-48
Identities = 61/89 (68%), Positives = 74/89 (82%)
Frame = +1
Query: 1 MLVFQARSLRRIEAFTQTCINRRSATVYSCYSDAPHKKSKLAPLQERRMIDKFKIIAKAG 60
MLVF+AR +R+++ FT+TC++R S YSCYS++PH KSKLAPLQERRMIDK KI AKAG
Sbjct: 79 MLVFEARYVRQVKVFTRTCVSRWSTVFYSCYSNSPHSKSKLAPLQERRMIDKVKIFAKAG 258
Query: 61 DGGSGCSSFHRSRHDRQGRADGGNGGRGG 89
DGG GC+S RSRHDR+ ADGG+GG GG
Sbjct: 259 DGGHGCTSSRRSRHDRKSIADGGSGGVGG 345
Score = 78.6 bits (192), Expect(2) = 2e-48
Identities = 42/64 (65%), Positives = 43/64 (66%)
Frame = +2
Query: 89 GDVILECSPRVWDFSGLQHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVIHLVNGDIP 148
GDVILECS RVWDFSGLQ HLIAG GG GSSKN IGTRG +GDIP
Sbjct: 344 GDVILECSRRVWDFSGLQRHLIAGNGGHGSSKNXIGTRGCR*GCSCTDWHCTSSCSGDIP 523
Query: 149 SIVK 152
SIVK
Sbjct: 524 SIVK 535
>TC91937 similar to GP|22136032|gb|AAM91598.1 GTP-binding protein obg-like
{Arabidopsis thaliana}, partial (45%)
Length = 1011
Score = 142 bits (358), Expect = 3e-34
Identities = 95/245 (38%), Positives = 131/245 (52%), Gaps = 5/245 (2%)
Frame = +3
Query: 252 VAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLNA 311
+ E+ GQ+ ++ GG GG GN + S K + + +
Sbjct: 291 ILEMVYPGQKALLLPGGRGGRGNAAFKSGSNK------------------APKIAEN--- 407
Query: 312 GMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNF 371
G G E L LELK +ADV VG PNAGKSTLL +S AKP V +Y FTTL PNLG ++F
Sbjct: 408 GEQGHEMWLELELKLVADVGIVGAPNAGKSTLLSVVSAAKPEVANYPFTTLLPNLGVVSF 587
Query: 372 D-DFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQL 430
D D ++ VAD+PGL++GAH+ GLGH FLRH ER L +VVD G P L
Sbjct: 588 DYDSTMVVADLPGLLEGAHRGFGLGHEFLRHTERCSALIHVVD---------GSSPQPDL 740
Query: 431 K-DLI-LELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRV--HGVPIFRVCAVLEE 486
+ D + LEL+ L+ +P ++ NK+D A E + + ++ HG+ F + AV E
Sbjct: 741 EFDAVRLELKLFNPELAEKPFIVAYNKMDLPEAYENWESFKEKLQSHGITPFCMSAVKRE 920
Query: 487 GIPEL 491
G E+
Sbjct: 921 GTHEV 935
Score = 68.9 bits (167), Expect = 4e-12
Identities = 37/86 (43%), Positives = 51/86 (59%), Gaps = 2/86 (2%)
Frame = +3
Query: 57 AKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQH--HLIAGKG 114
AKAGDGG+G +F R ++ G GG+GGRGG+V +E + ++ H AG+G
Sbjct: 9 AKAGDGGNGSMAFRREKYVPFGGPSGGDGGRGGNVYVEVDKAMNSLLPFRNGIHFRAGRG 188
Query: 115 GPGSSKNKIGTRGADKVVHVPVGTVI 140
G + +IG +G D VV VP GTVI
Sbjct: 189 SHGQGRMQIGAKGDDVVVKVPPGTVI 266
>TC86616 similar to GP|7643796|gb|AAF65513.1| GTP-binding protein {Capsicum
annuum}, partial (57%)
Length = 886
Score = 60.8 bits (146), Expect = 1e-09
Identities = 33/113 (29%), Positives = 54/113 (47%), Gaps = 17/113 (15%)
Frame = +2
Query: 317 ENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDDFS- 375
E ++ S + VG+PN GKSTL +++ ++ F T+ PN +N D
Sbjct: 146 ERPILGRFSSHLKIGIVGLPNVGKSTLFNTLTKMAIPAENFPFCTIEPNEARVNVPDERF 325
Query: 376 ----------------ITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVV 412
+ + DI GL++GAHQ +GLG++FL HI + +V+
Sbjct: 326 EWLCQLFKPKSEVSAFLEIHDIAGLVRGAHQGQGLGNSFLSHIRAVDGIFHVL 484
>CB891834 homologue to PIR|C96751|C96 probable GTP-binding protein F28P22.15
[imported] - Arabidopsis thaliana, partial (43%)
Length = 518
Score = 58.5 bits (140), Expect = 6e-09
Identities = 39/122 (31%), Positives = 62/122 (49%)
Frame = +3
Query: 330 VSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDDFSITVADIPGLIKGAH 389
VS +G P+ GKSTLL ++ Y FTTL G ++++D I + D+PG+I+GA
Sbjct: 174 VSLIGFPSVGKSTLLTLLTGTHSEAASYEFTTLTCIPGIIHYNDTKIQLLDLPGIIEGAS 353
Query: 390 QNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQLKDLILELEYHQDGLSNRPS 449
+ +G G + + + ++ V+D A+ G + I L ELE L+ RP
Sbjct: 354 EGKGRGRQVIAVAKSSDIVLMVLD-ASKSEGHRQI--------LTKELEAVGLRLNKRPP 506
Query: 450 LI 451
I
Sbjct: 507 QI 512
>BG644931 similar to GP|22135878|gb GTP-binding protein putative
{Arabidopsis thaliana}, partial (47%)
Length = 697
Score = 53.5 bits (127), Expect = 2e-07
Identities = 53/173 (30%), Positives = 81/173 (46%), Gaps = 13/173 (7%)
Frame = +3
Query: 333 VGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDDFSITVADIPGLIKGAHQNR 392
VG PN GKS+L+ +S KP + +Y FTT +G++ F+ V D PGL+K +R
Sbjct: 186 VGAPNVGKSSLVHVLSTGKPEICNYPFTTRGILMGHIVFNHQKFQVTDTPGLLKRHDDDR 365
Query: 393 G----LGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQLKDLILELEYHQDG----- 443
L A L ++ T VL YV DL+ G G P +Q + E++ DG
Sbjct: 366 NNLEKLTLAVLSYLP-TAVL-YVHDLS----GECGTSPSDQY-SIYKEIKERFDGHLWLD 524
Query: 444 LSNRPSLIVANKI----DEEGADEVYLELQRRVHGVPIFRVCAVLEEGIPELK 492
+ ++ L+ + + ++ + LE R+ V EEGI ELK
Sbjct: 525 VVSKADLLRTSPVIYATEDRDLTQHELEKYRKSGPDGAINVSVTTEEGIHELK 683
>TC88185 homologue to PIR|T09368|T09368 GTP-binding protein F23K16.150 -
Arabidopsis thaliana, complete
Length = 1734
Score = 52.8 bits (125), Expect = 3e-07
Identities = 29/86 (33%), Positives = 42/86 (48%)
Frame = +3
Query: 328 ADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDDFSITVADIPGLIKG 387
A V VG P+ GKSTLL ++ V Y FTTL G + + I + D+PG+I+G
Sbjct: 342 ARVGLVGFPSVGKSTLLNKLTGTFSEVASYEFTTLTCIPGVITYRGAKIQLLDLPGIIEG 521
Query: 388 AHQNRGLGHAFLRHIERTKVLAYVVD 413
A +G G + + V+D
Sbjct: 522 AKDGKGRGRQVISTARTCNCILIVLD 599
>TC88979 similar to PIR|H96601|H96601 hypothetical protein T6H22.14
[imported] - Arabidopsis thaliana, partial (89%)
Length = 1616
Score = 45.1 bits (105), Expect = 7e-05
Identities = 28/79 (35%), Positives = 36/79 (45%), Gaps = 18/79 (22%)
Frame = +3
Query: 333 VGMPNAGKSTLLGA-ISRAKPAVGHYAFTTLRPNLGNLNFDD-----------------F 374
VG+PN GKSTL A + K ++ F T+ PN+G + D
Sbjct: 219 VGLPNVGKSTLFNAVVENGKAQAANFPFCTIEPNVGIVAVPDSRLQVLSDLSKSQRMVPA 398
Query: 375 SITVADIPGLIKGAHQNRG 393
SI DI GL+KGA RG
Sbjct: 399 SIEFVDIAGLVKGASSRRG 455
>BF640447 similar to GP|20152085|gb| RE21802p {Drosophila melanogaster},
partial (15%)
Length = 326
Score = 36.2 bits (82), Expect = 0.031
Identities = 14/36 (38%), Positives = 23/36 (63%)
Frame = +2
Query: 330 VSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPN 365
V VG+PN GKST +++++ A ++ F T+ PN
Sbjct: 215 VGIVGVPNVGKSTFFNVLTKSQAAAENFPFCTIDPN 322
>TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsis thaliana},
partial (31%)
Length = 2756
Score = 32.0 bits (71), Expect = 0.59
Identities = 17/34 (50%), Positives = 23/34 (67%)
Frame = +1
Query: 195 TGCSSSQAAEKNVEKSVKSGHVASTDVLSQLSIS 228
TGCSSS ++ K+ S+ SG V STD ++ SIS
Sbjct: 1291 TGCSSSTSSSKSSSNSI-SGTVTSTDTVTSSSIS 1389
>TC78399 homologue to SP|O04834|SARA_ARATH GTP-binding protein SAR1A.
[Mouse-ear cress] {Arabidopsis thaliana}, complete
Length = 1164
Score = 31.2 bits (69), Expect = 1.0
Identities = 34/133 (25%), Positives = 58/133 (43%), Gaps = 3/133 (2%)
Frame = +2
Query: 328 ADVSFVGMPNAGKSTLLGAISRAKPAVGHYAF---TTLRPNLGNLNFDDFSITVADIPGL 384
A + F+G+ NAGK+TLL + + + V H T+ ++G + F F +
Sbjct: 134 AKILFLGLDNAGKTTLLHML-KDERLVQHQPTQHPTSEELSIGRIKFKAFDL-------- 286
Query: 385 IKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQLKDLILELEYHQDGL 444
G HQ + + + + Y+VD ++ P ++ D +L + L
Sbjct: 287 --GGHQI--ARRVWRDYYAQVDAVVYLVD----AFDKERFPESKKELDALLA----DESL 430
Query: 445 SNRPSLIVANKID 457
N P LI+ NKID
Sbjct: 431 GNVPFLILGNKID 469
>TC83590 similar to GP|9758365|dbj|BAB08866.1 GTP binding protein-like
{Arabidopsis thaliana}, partial (49%)
Length = 1061
Score = 30.8 bits (68), Expect = 1.3
Identities = 17/35 (48%), Positives = 19/35 (53%)
Frame = +2
Query: 330 VSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRP 364
VS VG NAGKSTLL ++ A F TL P
Sbjct: 884 VSLVGYTNAGKSTLLNQLTGADVLAEDKLFATLDP 988
>BQ141699 similar to GP|334068|gb|A ORF2 {Pseudorabies virus}, partial (1%)
Length = 1217
Score = 30.8 bits (68), Expect = 1.3
Identities = 15/41 (36%), Positives = 20/41 (48%)
Frame = +3
Query: 55 IIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILEC 95
++ K+ GGSGC R R D R G + G GD +C
Sbjct: 744 VLRKSARGGSGCRE*RRLRSDASARRVGRSQGARGDTDDKC 866
>TC85599 homologue to SP|O04834|SARA_ARATH GTP-binding protein SAR1A.
[Mouse-ear cress] {Arabidopsis thaliana}, complete
Length = 999
Score = 30.4 bits (67), Expect = 1.7
Identities = 33/133 (24%), Positives = 59/133 (43%), Gaps = 3/133 (2%)
Frame = +2
Query: 328 ADVSFVGMPNAGKSTLLGAISRAKPAVGHYAF---TTLRPNLGNLNFDDFSITVADIPGL 384
A + F+G+ NAGK+TLL + + + V H T+ ++G + F F +
Sbjct: 215 AKILFLGLDNAGKTTLLHML-KDERLVQHQPTQHPTSEELSIGKIKFKAFDL-------- 367
Query: 385 IKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQLKDLILELEYHQDGL 444
G HQ + + + + Y+VD + ++ ++ D +L + L
Sbjct: 368 --GGHQI--ARRVWKDYYAKVDAVVYLVD----AYDKERFAESKKELDALLS----DESL 511
Query: 445 SNRPSLIVANKID 457
+N P LI+ NKID
Sbjct: 512 ANVPFLILGNKID 550
>TC85598 homologue to SP|O04834|SARA_ARATH GTP-binding protein SAR1A.
[Mouse-ear cress] {Arabidopsis thaliana}, complete
Length = 900
Score = 29.6 bits (65), Expect = 2.9
Identities = 32/133 (24%), Positives = 59/133 (44%), Gaps = 3/133 (2%)
Frame = +3
Query: 328 ADVSFVGMPNAGKSTLLGAISRAKPAVGHYAF---TTLRPNLGNLNFDDFSITVADIPGL 384
A + F+G+ NAGK+TLL + + + V H T+ ++G + F F +
Sbjct: 234 AKILFLGLDNAGKTTLLHML-KDERLVQHQPTQYPTSEELSIGKIKFKAFDL-------- 386
Query: 385 IKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQLKDLILELEYHQDGL 444
G HQ + + + + Y+VD + ++ ++ D +L + L
Sbjct: 387 --GGHQI--ARRVWKDYYAQVDAVVYLVD----AYDKERFAESKKELDALLS----DESL 530
Query: 445 SNRPSLIVANKID 457
+N P L++ NKID
Sbjct: 531 ANVPFLVLGNKID 569
>BI272296 similar to GP|12322014|gb GTPase putative; 34281-30152
{Arabidopsis thaliana}, partial (22%)
Length = 487
Score = 29.3 bits (64), Expect = 3.8
Identities = 18/45 (40%), Positives = 26/45 (57%)
Frame = +2
Query: 303 DSVCSSLNAGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAI 347
D VCS L + +N L+ E + +S VG PN GKS++L A+
Sbjct: 107 DLVCSGLXK-VEEPQN-LVEEEDYVPAISIVGRPNVGKSSILNAL 235
>AW559547 weakly similar to GP|12654451|gb| ADP-ribosylation factor-like 7
{Homo sapiens}, partial (22%)
Length = 394
Score = 28.9 bits (63), Expect = 5.0
Identities = 38/135 (28%), Positives = 61/135 (45%), Gaps = 6/135 (4%)
Frame = +2
Query: 329 DVSFVGMPNAGKSTLLGAISRAKPAVGHYAFT--TLRPNLGNLNFDDFSITVADIPGLIK 386
+V VG+ N+GK+TLL + A+GH T T+ N+ + + DI G +
Sbjct: 62 EVVLVGLENSGKTTLLNVM-----AMGHPVETCPTIGLNVKLVKKGGVQMKCWDIGGQAQ 226
Query: 387 GAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQL----KDLILELEYHQD 442
+ + R+ V+ YVVD A ++Q+ K+L LE +
Sbjct: 227 YRSE-------WGRYTRGCDVIIYVVDANA----------FDQISLARKELHRLLEDRE- 352
Query: 443 GLSNRPSLIVANKID 457
L+ P L++ANKID
Sbjct: 353 -LATTPLLVLANKID 394
>BQ136093
Length = 838
Score = 28.9 bits (63), Expect = 5.0
Identities = 14/24 (58%), Positives = 17/24 (70%), Gaps = 1/24 (4%)
Frame = -1
Query: 72 SRH-DRQGRADGGNGGRGGDVILE 94
SRH + G+ GNGGR G+VILE
Sbjct: 559 SRHAESDGKRGRGNGGRAGNVILE 488
>TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {Arabidopsis
thaliana}, partial (92%)
Length = 1250
Score = 28.5 bits (62), Expect = 6.5
Identities = 24/79 (30%), Positives = 29/79 (36%), Gaps = 3/79 (3%)
Frame = +3
Query: 60 GDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQHHLIAGKGGPGSS 119
G GG G +G GG GGRGGD R G G+GG G
Sbjct: 75 GRGGGGFRGGRGGDRGGRGGGRGGFGGRGGDRGTPFKARGGGRGG------GGRGGRGGG 236
Query: 120 K---NKIGTRGADKVVHVP 135
+ G +G KV+ P
Sbjct: 237 RGGGRGGGMKGGSKVIVEP 293
>TC81771 similar to GP|10176714|dbj|BAB09944. dimethylaniline
monooxygenase-like protein {Arabidopsis thaliana},
partial (39%)
Length = 874
Score = 28.5 bits (62), Expect = 6.5
Identities = 14/41 (34%), Positives = 24/41 (58%)
Frame = +2
Query: 414 LAAALHGRKGIPPWEQLKDLILELEYHQDGLSNRPSLIVAN 454
+A L G+K +P WE++ I E YH+ ++ P+ +AN
Sbjct: 272 IAQLLSGKKVLPSWEEMMKSIEEF-YHEREVAGIPTHDIAN 391
>TC81923 GP|21667854|gb|AAM74155.1 cyclin T1 protein {Capra hircus}, partial
(1%)
Length = 839
Score = 28.1 bits (61), Expect = 8.5
Identities = 15/36 (41%), Positives = 19/36 (52%)
Frame = -1
Query: 82 GGNGGRGGDVILECSPRVWDFSGLQHHLIAGKGGPG 117
GG GG GG ++ V+ G H + GKGGPG
Sbjct: 809 GGYGGGGG--VMMHQGHVYGSVGGGFHQVMGKGGPG 708
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,241,706
Number of Sequences: 36976
Number of extensions: 185647
Number of successful extensions: 701
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 685
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 693
length of query: 515
length of database: 9,014,727
effective HSP length: 100
effective length of query: 415
effective length of database: 5,317,127
effective search space: 2206607705
effective search space used: 2206607705
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0176a.13


