

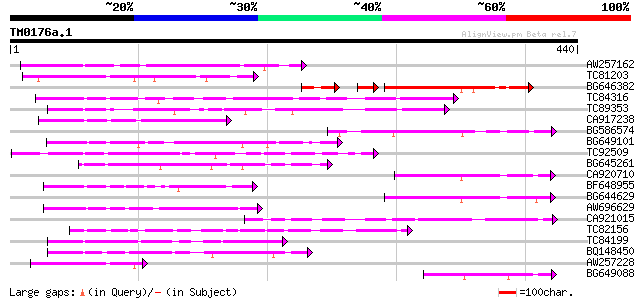
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0176a.1
(440 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW257162 140 7e-34
TC81203 weakly similar to GP|10177464|dbj|BAB10855. gb|AAD21700.... 122 4e-28
BG646382 100 6e-27
TC84316 weakly similar to GP|11994612|dbj|BAB02749. gb|AAC24186.... 111 5e-25
TC89353 weakly similar to GP|9279704|dbj|BAB01261.1 gb|AAF25964.... 108 4e-24
CA917238 weakly similar to PIR|A84538|A845 hypothetical protein ... 107 9e-24
BG586574 104 6e-23
BG649101 homologue to GP|6648199|gb|A unknown protein {Arabidops... 104 6e-23
TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein... 97 2e-20
BG645261 homologue to PIR|H90254|H902 sulfate ABC transporter p... 95 5e-20
CA920710 similar to PIR|S64314|S643 probable membrane protein YG... 92 5e-19
BF648955 similar to GP|18043667|gb| Unknown (protein for IMAGE:3... 87 2e-17
BG644629 86 2e-17
AW696629 84 1e-16
CA921015 homologue to SP|Q8Y7G1|DPO3 DNA polymerase III polC-typ... 76 3e-14
TC82156 similar to GP|20197985|gb|AAM15340.1 F-box protein famil... 74 9e-14
TC84199 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 72 4e-13
BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.1... 72 6e-13
AW257228 GP|17342711|gb 1-aminocyclopropanecarboxylic acid oxida... 71 9e-13
BG649088 70 2e-12
>AW257162
Length = 728
Score = 140 bits (354), Expect = 7e-34
Identities = 89/226 (39%), Positives = 131/226 (57%), Gaps = 4/226 (1%)
Frame = +1
Query: 9 VTVLSMNTPHPDNLPRTQLH-NDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKL 67
+T + MNT +L + + +D+IAE+LSWL VK+L ++KCV KSWN+L ISD F+K+
Sbjct: 70 ITAVVMNTRSRMSLSSSPVFPDDIIAEILSWLTVKTLMKMKCVSKSWNTL-ISDSNFVKM 246
Query: 68 HLHRSALHGKPHLLLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDR 127
HL+RSA H + +L+ + H F + G L+ I+L +P Y+L + D
Sbjct: 247 HLNRSARHSQSYLV----SEHRGDYNFVPFSVRG----LMNGRSITLPKDPYYQLIEKDC 402
Query: 128 YHVVGCCNGLICLHGYSVD-SIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGY 186
VVG CNGL+CL G D F++ WL WNPATR +S+KL + + ++ FGY
Sbjct: 403 PGVVGSCNGLVCLSGCVADVEEFEEMWLRIWNPATRTISDKLYFSANRLQCWE--FMFGY 576
Query: 187 DNSTDTYKVV--FFEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFP 230
DN+T TYKVV + + E + ++ C N WR+IQ+FP
Sbjct: 577 DNTTQTYKVVALYPDSEMTTKVGII-------CFRNNIWRNIQSFP 693
>TC81203 weakly similar to GP|10177464|dbj|BAB10855.
gb|AAD21700.1~gene_id:MQB2.18~similar to unknown protein
{Arabidopsis thaliana}, partial (9%)
Length = 715
Score = 122 bits (305), Expect = 4e-28
Identities = 78/199 (39%), Positives = 111/199 (55%), Gaps = 16/199 (8%)
Frame = +1
Query: 11 VLSMNTPHPDN--LPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLH 68
++S + P N P + L ++LI ++LS LPVK+L + KCVCKSW +LI D +F KLH
Sbjct: 145 LMSNSDPPQSNGGAPTSLLLDELIVDILSRLPVKTLMQFKCVCKSWKTLISHDPSFAKLH 324
Query: 69 LHRSALHGKPHLLLQRENSHHDHREFC-----VTHLSGSSLSLVE-NPW-----ISLAHN 117
L RS + HL L + S D F V+HL + L++ P+ +++ +
Sbjct: 325 LQRSPRN--THLTLVSDRSSDDESNFSVVPFPVSHLIEAPLTITPFEPYHLLRNVAIPDD 498
Query: 118 PRYRLKKGDRYHVVGCCNGLICLHGYSVDSIFQQ---GWLSFWNPATRLMSEKLGCFCVK 174
P Y L D ++G CNGL CL YS+ I+++ W FWNPAT +SE+LGC
Sbjct: 499 PYYVLGNMDCCIIIGSCNGLYCLRCYSL--IYEEEEHDWFRFWNPATNTLSEELGCL--- 663
Query: 175 TKNFDVKLSFGYDNSTDTY 193
N +L+FGYD S DTY
Sbjct: 664 --NEFFRLTFGYDISNDTY 714
>BG646382
Length = 603
Score = 100 bits (248), Expect(3) = 6e-27
Identities = 57/124 (45%), Positives = 78/124 (61%), Gaps = 9/124 (7%)
Frame = +3
Query: 292 PKGLDEVPSVVPSVGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKVRYQDLQ-- 349
P G EVP V PS+ VLM+CLCFSHD + T FVIWQMK+FG +SWTQL +++Y +LQ
Sbjct: 198 PAGFKEVPCVEPSLRVLMDCLCFSHDFKRTEFVIWQMKEFGSQESWTQLFRIKYINLQIH 377
Query: 350 ----REPLHSMFY--CHYQLFPLCL-HDGDTLILASYSDRYDGNEAILYDKRANTAERTA 402
+ L + Y C+ L PL L +GDTLIL+ +D +G I+Y++R E+T
Sbjct: 378 NLPINDNLDLLGYMECNIPLLPLYLSENGDTLILS--NDEEEG--VIIYNQRDKRVEKTR 545
Query: 403 LETE 406
+ E
Sbjct: 546 ISNE 557
Score = 31.6 bits (70), Expect(3) = 6e-27
Identities = 16/30 (53%), Positives = 20/30 (66%)
Frame = +1
Query: 227 QNFPLLPFSYDDWGVNDGVHLSGTINWMTS 256
Q+FPL+P + NDGVHLSG IN + S
Sbjct: 1 QSFPLVPLIW-----NDGVHLSGAINLVGS 75
Score = 27.3 bits (59), Expect(3) = 6e-27
Identities = 13/16 (81%), Positives = 14/16 (87%)
Frame = +2
Query: 271 VEQLGILSLDLRTETY 286
VEQL I+SLDL TETY
Sbjct: 134 VEQLVIVSLDLSTETY 181
>TC84316 weakly similar to GP|11994612|dbj|BAB02749.
gb|AAC24186.1~gene_id:MGL6.4~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (10%)
Length = 892
Score = 111 bits (278), Expect = 5e-25
Identities = 97/331 (29%), Positives = 150/331 (45%), Gaps = 3/331 (0%)
Frame = +3
Query: 21 NLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHL 80
N P L +LI +L LPV+SL R KCVCKSW +L SD F H S ++ P L
Sbjct: 24 NKPLPFLPEELIVIILLRLPVRSLLRFKCVCKSWKTLF-SDTHFANNHFLISTVY--PQL 194
Query: 81 LLQRENSHHDHREFCVTHLSGSSLSLVENPWISL--AHNPRYRLKKGDRYHVVGCCNGLI 138
+ S + E + SL+EN ++ N ++ RY ++G CNG +
Sbjct: 195 VACESVSAYRTWEIKTYPIE----SLLENSSTTVIPVSNTGHQ-----RYTILGSCNGFL 347
Query: 139 CLHGYSVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFF 198
CL+ Q + WNP+ L S+ T + + FGYD YK++
Sbjct: 348 CLYDN------YQRCVRLWNPSINLKSKSS-----PTIDRFIYYGFGYDQVNHKYKLLAV 494
Query: 199 EIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINWMTS-R 257
+ +++ F + C ++++FP P + G +SGT+NW+ R
Sbjct: 495 KAFSRITETMIYTFGENSCKNV----EVKDFPRYPPNRKHLGK----FVSGTLNWIVDER 650
Query: 258 DKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVLMNCLCFSHD 317
D A ILS D+ ETY + P+ V S P + VL NC+C
Sbjct: 651 DGRAT-------------ILSFDIEKETYRQVLLPQHGYAVYS--PGLYVLSNCICVCTS 785
Query: 318 LEGTHFVIWQMKKFGDAKSWTQLLKVRYQDL 348
T + +W MKK+G A+SWT+L+ + +++L
Sbjct: 786 FLDTRWQLWMMKKYGVAESWTKLMSIPHENL 878
>TC89353 weakly similar to GP|9279704|dbj|BAB01261.1
gb|AAF25964.1~gene_id:MYA6.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1482
Score = 108 bits (270), Expect = 4e-24
Identities = 99/331 (29%), Positives = 147/331 (43%), Gaps = 19/331 (5%)
Frame = +3
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHH 89
++I E+L LPV+SL + +CVCK W +L ISD F K H+ S + P L+
Sbjct: 114 EIIVEILLRLPVRSLLQFRCVCKLWKTL-ISDPQFAKKHVSISTAY--PQLV-------- 260
Query: 90 DHREFCVTHLSGSSLSLVENPWISLAHNP-RYRLKKGD-------RYHVVGCCNGLICLH 141
+S + +LV P L NP +R++ D ++G CNGL+CL
Sbjct: 261 ------SVFVSIAKCNLVSYPLKPLLDNPSAHRVEPADFEMIHTTSMTIIGSCNGLLCL- 419
Query: 142 GYSVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVK----LSFGYDNSTDTYKVVF 197
S F Q + WNP+ +L S K + +FD K FGYD D YKV+
Sbjct: 420 -----SDFYQ--FTLWNPSIKLKS-KPSPTIIAFDSFDSKRFLYRGFGYDQVNDRYKVL- 572
Query: 198 FEIEKIRRASLVSVFTLDGCN------GWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTI 251
A + + + LD G W IQ FP P D + G +SG +
Sbjct: 573 --------AVVQNCYNLDETKTLIYTFGGKDWTTIQKFPCDPSRCDLGRLGVGKFVSGNL 728
Query: 252 NWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVLMNC 311
NW+ S+ I+ D+ ETY +S P+ + +V + V N
Sbjct: 729 NWIVSKKV----------------IVFFDIEKETYGEMSLPQDYGDKNTV---LYVSSNR 851
Query: 312 LCFSHDLEG-THFVIWQMKKFGDAKSWTQLL 341
+ S D TH+V+W MK++G +SWT+L+
Sbjct: 852 IYVSFDHSNKTHWVVWMMKEYGVVESWTKLM 944
>CA917238 weakly similar to PIR|A84538|A845 hypothetical protein At2g16220
[imported] - Arabidopsis thaliana, partial (6%)
Length = 603
Score = 107 bits (267), Expect = 9e-24
Identities = 64/150 (42%), Positives = 87/150 (57%)
Frame = +2
Query: 23 PRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLL 82
P + L ++LI EVLS VKSL R++CV + WNS+I SD F+KLH+ RSA + HL L
Sbjct: 134 PPSVLPDELITEVLSRGDVKSLMRMRCVSEYWNSMI-SDPRFVKLHMKRSARNA--HLTL 304
Query: 83 QRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHG 142
S D V + L+EN I+L +P YRLK + +V+G CNG +CL G
Sbjct: 305 SLCKSGIDGDNNVVPY---PVRGLIENGLITLPSDPYYRLKDKECQYVIGSCNGWLCLLG 475
Query: 143 YSVDSIFQQGWLSFWNPATRLMSEKLGCFC 172
+S ++ W FWNPA M++KLG C
Sbjct: 476 FSSIGAYRHIWFRFWNPAMGKMTQKLGYIC 565
>BG586574
Length = 732
Score = 104 bits (260), Expect = 6e-23
Identities = 79/189 (41%), Positives = 98/189 (51%), Gaps = 11/189 (5%)
Frame = +1
Query: 247 LSGTINWM--TSRDKSAPYYNPVTITVEQLGILSLDLRTETYT-LLSPPKGLD------E 297
L+ T+NW+ AP QL I+SLDL TETYT L PP LD +
Sbjct: 1 LNSTLNWLGFIQDGDLAP----------QLVIISLDLGTETYTQFLPPPISLDLSHVLQK 150
Query: 298 VPSVVPSVGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKVRYQDLQR--EPLHS 355
V P V +LM+ LCF HDL T FVIW+M KFGD KSW QLLK+ Y L+ +P S
Sbjct: 151 VSHAKPGVSMLMDSLCFYHDLNETDFVIWKMTKFGDEKSWAQLLKISYHKLKMNLKPGIS 330
Query: 356 MFYCHYQLFPLCLHDGDTLILASYSDRYDGNEAILYDKRANTAERTALETESKYILGFSS 415
MF + +GDTL+ AILY+ R N +T + +K I FS
Sbjct: 331 MFNLYV--------NGDTLVFVDDQK----ERAILYNWRNNRVVKTRV---NKKICWFSI 465
Query: 416 FYYVESLVS 424
+YVESLVS
Sbjct: 466 NHYVESLVS 492
>BG649101 homologue to GP|6648199|gb|A unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 799
Score = 104 bits (260), Expect = 6e-23
Identities = 84/239 (35%), Positives = 127/239 (52%), Gaps = 9/239 (3%)
Frame = +1
Query: 29 NDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSH 88
+DLIA +L++LPVK++T+LK V KSWN+LI S +FIK+HL++S+ + P+ +L
Sbjct: 91 SDLIAIILTFLPVKTITQLKLVSKSWNTLITS-PSFIKIHLNQSSQN--PNFILTPSRKQ 261
Query: 89 HDHREFCVTH----LSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYS 144
+ L+G+++S + + ++ +N + + VVG CNGL+CL S
Sbjct: 262 YSINNVLSVPIPRLLTGNTVS--GDTYHNILNNDHH-------FRVVGSCNGLLCLLFKS 414
Query: 145 VDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFD--VKLSFGYDNSTDTYKVVFF---E 199
+ WNPATR +SE+LG F F + +FG D TYK+V E
Sbjct: 415 EFITHLKFRFRIWNPATRTISEELGFFRKYKPLFGGVSRFTFGCDYLRGTYKLVALHTVE 594
Query: 200 IEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINWMTSRD 258
+ R++ V VF L + WR+I N PF DGVHLSGT NW++ R+
Sbjct: 595 DGDVMRSN-VRVFNLGNDDSDKCWRNIPN----PFV-----CADGVHLSGTGNWLSLRE 741
>TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein F26G5.80 -
Arabidopsis thaliana, partial (8%)
Length = 858
Score = 96.7 bits (239), Expect = 2e-20
Identities = 95/291 (32%), Positives = 132/291 (44%), Gaps = 6/291 (2%)
Frame = +3
Query: 2 LPNSNNTVTVLSMNTPHPDNLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISD 61
+ S T+T S + P LP +L+AE+L LPVK L +L+C+CKS+NSL ISD
Sbjct: 114 MQQSTETLTPQSRHAPPLPTLPF-----ELVAEILCRLPVKLLXQLRCLCKSFNSL-ISD 275
Query: 62 QTFIKLHLHRSALHGKPHLLLQRENSHHDHREFCVTHL-SGSSLSLVENPWISLAHNPRY 120
F K HLH S PH L+ R N+ V+ + S S S V P L +
Sbjct: 276 PKFAKKHLHSST---TPHHLILRSNNGSGRFALIVSPIQSVLSTSTVPVPQTQLTYPTCL 446
Query: 121 RLKKGDRYHVVGCCNGLICLHGYSVDSIFQQGWLSFWN-----PATRLMSEKLGCFCVKT 175
+ Y C+G+ICL + D W F N P + +S K C+ T
Sbjct: 447 TEEFASPYEWCS-CDGIICL---TTDYSSAVLWNPFINKFKTLPPLKYISLKRSPSCLFT 614
Query: 176 KNFDVKLSFGYDNSTDTYKVVFFEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFS 235
FGYD D YKV F I + + V V T+ G + WR I++FP F
Sbjct: 615 --------FGYDPFADNYKV--FAITFCVKRTTVEVHTM----GTSSWRRIEDFPSWSFI 752
Query: 236 YDDWGVNDGVHLSGTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETY 286
D G+ ++G ++W+T Y P + Q I+SLDL E+Y
Sbjct: 753 PD-----SGIFVAGYVHWLT-------YDGPGS----QREIVSLDLEDESY 857
>BG645261 homologue to PIR|H90254|H902 sulfate ABC transporter permease
protein SSO1032 [imported] - Sulfolobus solfataricus,
partial (4%)
Length = 771
Score = 95.1 bits (235), Expect = 5e-20
Identities = 77/212 (36%), Positives = 108/212 (50%), Gaps = 15/212 (7%)
Frame = +1
Query: 54 WNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHHDHREFCVTHLSGSSLSLVENPWIS 113
W S II TF+KLHL+RS+ KP L +S + L+ENP +
Sbjct: 130 WKS-IIDHPTFVKLHLNRSSQ--KPDLTFVSSARITLWTLESTCTVSFTVFRLLENPPVI 300
Query: 114 LA---HNPRYRLKKGDRYHVVGCCNGLICLHGYSVDSIFQQGWLS--FWNPATRLMSEKL 168
+ +P Y+LK D H++G CNGL+CL G ++ +S FWNPA+R +S+K+
Sbjct: 301 INLSDDDPYYQLKDKDCVHIIGSCNGLLCLFGLKINDSCGHMDISIRFWNPASRKISKKV 480
Query: 169 GCFCVKTKNFD----------VKLSFGYDNSTDTYKVVFFEIEKIRRASLVSVFTLDGCN 218
G +C N D +K FGY NS+DTYKVV+F I + VF+L
Sbjct: 481 G-YCGDGVNNDPSCFPGKGDLLKFVFGYVNSSDTYKVVYF----IVGTTSARVFSL---- 633
Query: 219 GWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGT 250
G N WR I+N P++ V+ VHLSG+
Sbjct: 634 GDNVWRKIENSPVVIHH-----VSGFVHLSGS 714
>CA920710 similar to PIR|S64314|S643 probable membrane protein YGR023w -
yeast (Saccharomyces cerevisiae), partial (7%)
Length = 774
Score = 91.7 bits (226), Expect = 5e-19
Identities = 54/128 (42%), Positives = 74/128 (57%), Gaps = 3/128 (2%)
Frame = -2
Query: 299 PSVVPSVGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKVRYQDLQ--REPLHSM 356
P P + VL++CLCF HDL T FVIWQMK+FG +SWT+L K+ Y DLQ P+
Sbjct: 773 PCAEPYLRVLLDCLCFLHDLGKTEFVIWQMKEFGVRESWTRLFKIPYVDLQMLNLPIDVQ 594
Query: 357 FYCHYQLFPLCL-HDGDTLILASYSDRYDGNEAILYDKRANTAERTALETESKYILGFSS 415
+ Y + PL + +GD +IL + D + I+Y+KR +R + E I FS+
Sbjct: 593 YLNEYPMLPLYISKNGDKVILTNEKD----DATIIYNKRDKRVDRARISNE---IHWFSA 435
Query: 416 FYYVESLV 423
YVESLV
Sbjct: 434 MDYVESLV 411
>BF648955 similar to GP|18043667|gb| Unknown (protein for IMAGE:3661715) {Mus
musculus}, partial (3%)
Length = 514
Score = 86.7 bits (213), Expect = 2e-17
Identities = 60/168 (35%), Positives = 92/168 (54%), Gaps = 2/168 (1%)
Frame = +3
Query: 27 LHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQREN 86
L ++LI E++SWLPVK L + +CV K + +LI SD F+++HL +SA + PHL L ++
Sbjct: 57 LPSELIVEIISWLPVKYLMQFRCVSKFYKTLI-SDPYFVQMHLEKSARN--PHLALMWQD 227
Query: 87 SHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYH--VVGCCNGLICLHGYS 144
+ + LS S L + N + + P ++ G YH ++G CNGL+CL +
Sbjct: 228 DLL-REDGSIIFLSVSRL--LGNKYTT----PPFQ--SGTFYHSCIIGSCNGLLCLVDFH 380
Query: 145 VDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDT 192
+ +L FWNPATR + + T + D K SFGYD + T
Sbjct: 381 CPDYYYYYYLYFWNPATRTKFXNI----LITLSXDFKFSFGYDTLSKT 512
>BG644629
Length = 546
Score = 86.3 bits (212), Expect = 2e-17
Identities = 56/148 (37%), Positives = 80/148 (53%), Gaps = 16/148 (10%)
Frame = -3
Query: 292 PKGLDEVPSVVPSVGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKVRYQDLQ-- 349
P+ E+PS +P V V LC S+ + T F+IWQMK+ + +SWTQ LK+RYQ+LQ
Sbjct: 532 PRDFHEMPSALPIVAV*GGFLCCSYFYKETDFLIWQMKELANDESWTQFLKIRYQNLQIN 353
Query: 350 ------REPLHSMFYCHYQLFPLCL-HDGDTLILASYSDRYDGNEAILYDKRANTAERTA 402
E +H H+QL PL L + DTL+L + + + ILY+ + N +RT
Sbjct: 352 HDYFSDHEFIHDTVKYHFQLVPLLLSEEADTLVLKTSQE----SHPILYNWKENRVQRTK 185
Query: 403 LETES-------KYILGFSSFYYVESLV 423
+ T + IL S+ YVESLV
Sbjct: 184 VTTSTTVTDDSRNSILWCSAKVYVESLV 101
>AW696629
Length = 663
Score = 83.6 bits (205), Expect = 1e-16
Identities = 61/171 (35%), Positives = 88/171 (50%), Gaps = 1/171 (0%)
Frame = +2
Query: 27 LHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQREN 86
L +DLI ++LSWLPVK L R V K W SLI+ D F KLHL +S + H++L +
Sbjct: 65 LPSDLIMQILSWLPVKLLIRFTSVSKHWKSLIL-DPNFAKLHLQKSPKN--THMILTALD 235
Query: 87 SHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLH-GYSV 145
D + VT SL L ++ + + Y +VG NGL+CL S+
Sbjct: 236 DEDD--TWVVTPYPVRSLLLEQSSF----SDEECCCFDYHSYFIVGSTNGLVCLAVEKSL 397
Query: 146 DSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVV 196
++ + ++ FWNP+ RL S+K + +L FGYD+ DTYK V
Sbjct: 398 ENRKYELFIKFWNPSLRLRSKKAPSLNIGLYG-TARLGFGYDDLNDTYKAV 547
>CA921015 homologue to SP|Q8Y7G1|DPO3 DNA polymerase III polC-type (EC
2.7.7.7) (PolIII). {Listeria monocytogenes}, partial
(0%)
Length = 824
Score = 75.9 bits (185), Expect = 3e-14
Identities = 65/243 (26%), Positives = 106/243 (42%)
Frame = -1
Query: 183 SFGYDNSTDTYKVVFFEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVN 242
+FGYD+ D +V+F + + VSV+TL G + W+ I++ P F
Sbjct: 824 NFGYDHFIDNIRVMF-----VLPKNEVSVYTL----GTDYWKRIEDIPYNIFE------- 693
Query: 243 DGVHLSGTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVV 302
+GV +SGT+NW+ S D ILSLD+ E+Y + P + + +
Sbjct: 692 EGVFVSGTVNWLASDDSF---------------ILSLDVEKESYQQVLLP----DTENDL 570
Query: 303 PSVGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKVRYQDLQREPLHSMFYCHYQ 362
+GVL NCLC +W M ++G+ +SWT+L V +++ Y
Sbjct: 569 WILGVLRNCLCIL-ATSNLFLDVWIMNEYGNQESWTKLYSVPNMQDHGLEAYTVLYS--- 402
Query: 363 LFPLCLHDGDTLILASYSDRYDGNEAILYDKRANTAERTALETESKYILGFSSFYYVESL 422
+ D L+L R D + ++YD + T + I +Y+ESL
Sbjct: 401 ------SEDDQLLLEFNEMRSDKVKLVVYDSKTGTLNIPEFQNNYDQI---CQNFYIESL 249
Query: 423 VSP 425
+SP
Sbjct: 248 ISP 240
>TC82156 similar to GP|20197985|gb|AAM15340.1 F-box protein family AtFBX9
{Arabidopsis thaliana}, partial (4%)
Length = 1136
Score = 74.3 bits (181), Expect = 9e-14
Identities = 80/267 (29%), Positives = 120/267 (43%), Gaps = 1/267 (0%)
Frame = +3
Query: 47 LKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHHDHREFCVTHLSGSSLSL 106
+K V KSW SLI SD F K +L S H LL + + + F LS SL
Sbjct: 456 VKSVSKSWKSLI-SDSNFTKKNLRVSTTS---HRLLFPKLTKGQYI-FNACTLS----SL 608
Query: 107 VENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSVDSIFQQGWLSFWNPATRLMSE 166
+ + A ++K D+ + G C+G++CL Q + WNP +
Sbjct: 609 ITTKGTATAMQHPLNIRKFDK--IRGSCHGILCLE-------LHQRFAILWNPFINKYAS 761
Query: 167 KLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFFEIEKIRRASLVSVFTLDGCNGWNGWRDI 226
L + N + FGYD+STD+YKV F I+ + + + + G WR I
Sbjct: 762 -LPPLEIPWSN-TIYSCFGYDHSTDSYKVAAF-IKWMPNSEIYKTYV--HTMGTTSWRMI 926
Query: 227 QNFPLLPFSYDDWGVNDGVHLSGTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETY 286
Q+FP P+ + G +S T NW+ +DK +V L ++SL L E+Y
Sbjct: 927 QDFPCTPY------LKSGKFVSWTFNWLAYKDK---------YSVSSLLVVSLHLENESY 1061
Query: 287 -TLLSPPKGLDEVPSVVPSVGVLMNCL 312
+L P G +V S+ S+ VL +CL
Sbjct: 1062GEILQPDYGSVDVLSI--SLWVLRDCL 1136
>TC84199 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 673
Score = 72.0 bits (175), Expect = 4e-13
Identities = 56/186 (30%), Positives = 81/186 (43%)
Frame = +1
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHH 89
+LI ++L WLPVKSL R KCVCKSW SL ISD F H +A H + L +
Sbjct: 118 ELIIQILLWLPVKSLLRFKCVCKSWFSL-ISDTHFANSHFQITAKHSRRVLFMLNHVPTT 294
Query: 90 DHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSVDSIF 149
+F H +++S + NP + P L C G I LH
Sbjct: 295 LSLDFEALHCD-NAVSEIPNPIPNFVEPPCDSLDTNS-----SSCRGFIFLH-------- 432
Query: 150 QQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFFEIEKIRRASLV 209
L WNP+TR+ +++ + +F FGYD D Y VV +++ +
Sbjct: 433 NDPDLFIWNPSTRVY-KQIPLSPNDSNSFHCLYGFGYDQLRDDYLVVSVTCQELMDYPCL 609
Query: 210 SVFTLD 215
F+ +
Sbjct: 610 RFFSFE 627
>BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.18~similar to
unknown protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 71.6 bits (174), Expect = 6e-13
Identities = 71/217 (32%), Positives = 101/217 (45%), Gaps = 11/217 (5%)
Frame = +3
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHH 89
++ E+LS LPVK L + +CVCK W S IS F+K HL S HL L +
Sbjct: 12 EIQVEILSRLPVKYLMQFQCVCKLWKSQ-ISKPDFVKKHLRVS---NTRHLFLLTFSKLS 179
Query: 90 DHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSVDSIF 149
E + SS+ P + P + D +VG C+G++C+
Sbjct: 180 P--ELVIKSYPLSSVFTEMTPTFTQLEYPLNNRDESD--SMVGSCHGILCI--------- 320
Query: 150 QQGWLSF---WNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVV-FFEIEKI-- 203
Q LSF WNP+ R ++ + K + +FGYD+S+DTYKVV F I
Sbjct: 321 -QCNLSFPVLWNPSIRKFTKLPSFEFPQNKFINPTYAFGYDHSSDTYKVVAVFCTSNIDN 497
Query: 204 ---RRASLVSVFTLDGCNGWNGWRDIQ-NFPL-LPFS 235
+ +LV+V T+ G N WR IQ FP +PF+
Sbjct: 498 GVYQLKTLVNVHTM----GTNCWRRIQTEFPFKIPFT 596
>AW257228 GP|17342711|gb 1-aminocyclopropanecarboxylic acid oxidase {Medicago
truncatula}, partial (11%)
Length = 702
Score = 70.9 bits (172), Expect = 9e-13
Identities = 43/98 (43%), Positives = 56/98 (56%), Gaps = 7/98 (7%)
Frame = +1
Query: 17 PHPD-NLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALH 75
PH + P + L ++LI E+LS LPVK+L + KCVCKSW +LI D F K HLHRS +
Sbjct: 313 PHSNFGAPTSLLLDELIVEILSRLPVKTLMQFKCVCKSWKTLISDDPVFAKFHLHRSPRN 492
Query: 76 GKPHLLLQRENSHHDHREFC------VTHLSGSSLSLV 107
HL + + S + C VTHL + LSLV
Sbjct: 493 --THLAILSDRSITEDETDCSVVPFPVTHLLEAPLSLV 600
>BG649088
Length = 683
Score = 69.7 bits (169), Expect = 2e-12
Identities = 49/110 (44%), Positives = 66/110 (59%), Gaps = 7/110 (6%)
Frame = -3
Query: 322 HFVIWQMKKFGDAKSWTQLLKVRYQDLQRE--PLHSMFY-CHYQLFPLCLHDGD-TLILA 377
HFV+W M ++G SWTQ LK+ +QDLQ + S+ Y L+P L + D TLI+A
Sbjct: 675 HFVLWLMMEYGVQDSWTQFLKISFQDLQIDYGISDSLDYGSQLYLYPFYLSESDNTLIMA 496
Query: 378 SYSDRYDG--NEAILYDKRANTAER-TALETESKYILGFSSFYYVESLVS 424
S YDG N AI+Y+ R T E+ T+++ E IL F + YVESLVS
Sbjct: 495 SNQQGYDGYDNHAIIYNWRDKTVEQITSVDNE---ILWFHTKDYVESLVS 355
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.137 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,360,573
Number of Sequences: 36976
Number of extensions: 328535
Number of successful extensions: 2020
Number of sequences better than 10.0: 119
Number of HSP's better than 10.0 without gapping: 1917
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1949
length of query: 440
length of database: 9,014,727
effective HSP length: 99
effective length of query: 341
effective length of database: 5,354,103
effective search space: 1825749123
effective search space used: 1825749123
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0176a.1


