

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
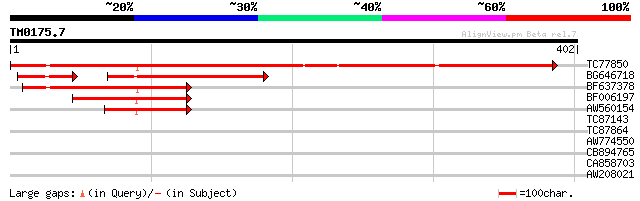
Query= TM0175.7
(402 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77850 weakly similar to PIR|H96517|H96517 protein T2E6.22 [imp... 421 e-118
BG646718 weakly similar to PIR|H96517|H96 protein T2E6.22 [impor... 121 3e-35
BF637378 weakly similar to PIR|H96517|H965 protein T2E6.22 [impo... 123 1e-28
BF006197 weakly similar to GP|1310677|emb| protein z-type serpin... 77 2e-14
AW560154 68 7e-12
TC87143 similar to GP|17976705|emb|CAC80839. dihydrofolate synth... 37 0.018
TC87864 similar to GP|16604637|gb|AAL24111.1 unknown protein {Ar... 30 2.2
AW774550 homologue to PIR|D84431|D84 probable endosomal protein ... 30 2.2
CB894765 29 3.7
CA858703 weakly similar to GP|22296462|db contains EST C25306(S5... 28 4.8
AW208021 similar to SP|O70422|TFH4_ TFIIH basal transcription fa... 28 8.2
>TC77850 weakly similar to PIR|H96517|H96517 protein T2E6.22 [imported] -
Arabidopsis thaliana, partial (65%)
Length = 1485
Score = 421 bits (1083), Expect = e-118
Identities = 229/392 (58%), Positives = 290/392 (73%), Gaps = 4/392 (1%)
Frame = +2
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
MDL++SI QT V+LS+ KHLFSKE + N+VFSP SL +LS++A GS G T +LF+
Sbjct: 41 MDLRESIANQTNVSLSVAKHLFSKES--DNNIVFSPLSLQVVLSIIASGSEGPTQQQLFN 214
Query: 61 FLRFDSVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTTHYK 118
FL+ S DHLN F SQ++S +LSD +P+ LLSFV+ +W D++L+L SF+Q+++TH+K
Sbjct: 215 FLQSKSTDHLNYFASQLVSVILSDASPAGGPLLSFVDGVWVDQTLSLQPSFQQIVSTHFK 394
Query: 119 ATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKH 178
A L+ VDF+ K +V EVNSW EKETNGLI ELLP +V+ T+LIFANALYFKG W
Sbjct: 395 AALSSVDFQNKAVEVTNEVNSWAEKETNGLIKELLPLGSVNNATRLIFANALYFKGAWND 574
Query: 179 TFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMC 238
FD S T +FHLLNG+ ++VPFM+S+KK QFIR FDGF VL L YKQG DK+ +F+M
Sbjct: 575 KFDASKTEDYEFHLLNGSPVKVPFMTSKKK-QFIRAFDGFKVLGLPYKQGEDKR-QFTMY 748
Query: 239 IFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGV 298
FLPNAKDGL +L+EK+ASES L+ KLP KV V FRIPKF I F LE S++LKELGV
Sbjct: 749 FFLPNAKDGLAALVEKVASESELLQHKLPFGKVEVGDFRIPKFNISFGLETSDMLKELGV 928
Query: 299 VSPFSKSNANFTKMVVSPLDE-LCVESIHHKASIEVNEEGTEAGVA-CSMCDVKLCVSAR 356
V PF S TKMV S + + LCV +I HK+ IEVNEEGTEA A + ++ +S
Sbjct: 929 VLPF--SGGGLTKMVNSSVSQNLCVSNIFHKSFIEVNEEGTEAAAATAATILLRSAMSIP 1102
Query: 357 TGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
+DFVADHPFLF+IREDLTGTI+F+GQVL+P
Sbjct: 1103PRLDFVADHPFLFMIREDLTGTIIFVGQVLNP 1198
>BG646718 weakly similar to PIR|H96517|H96 protein T2E6.22 [imported] -
Arabidopsis thaliana, partial (30%)
Length = 702
Score = 121 bits (304), Expect(2) = 3e-35
Identities = 65/114 (57%), Positives = 78/114 (68%)
Frame = +2
Query: 70 LNTFFSQVLSTVLSDTTPSFLLSFVNEMWADKSLALSHSFKQLMTTHYKATLALVDFRTK 129
L FS VLS S P LSFV+ +W +K+L+L SFKQ+M++ YKATLA VDF K
Sbjct: 209 LTACFSIVLSDAASAGGPH--LSFVDGVWIEKTLSLQPSFKQIMSSDYKATLASVDFMFK 382
Query: 130 GDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKHTFDPS 183
+V +EVN WVEKETNGLI EL+P +V LT LIFANALYF+G W FD S
Sbjct: 383 APEVRKEVNIWVEKETNGLIKELIPPGSVDSLTSLIFANALYFRGAWNEKFDVS 544
Score = 45.1 bits (105), Expect(2) = 3e-35
Identities = 23/43 (53%), Positives = 30/43 (69%)
Frame = +1
Query: 6 SIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAV 48
S+ QT V+L I KHLFSK+ E N+VFSP SL +LS++ V
Sbjct: 22 SMTNQTRVSLRIAKHLFSKQ--SENNIVFSPLSLQVVLSILIV 144
Score = 30.4 bits (67), Expect = 1.3
Identities = 21/48 (43%), Positives = 26/48 (53%), Gaps = 1/48 (2%)
Frame = +3
Query: 39 LHAILSVM-AVGSAGSTLDELFSFLRFDSVDHLNTFFSQVLSTVLSDT 85
L AI+S A S T +L FLR S DHLN SQ++S + S T
Sbjct: 99 LFAIVSASCAQHSDCPTPQQLLDFLRSKSTDHLNYLASQLVSLLCSPT 242
>BF637378 weakly similar to PIR|H96517|H965 protein T2E6.22 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 627
Score = 123 bits (308), Expect = 1e-28
Identities = 66/122 (54%), Positives = 86/122 (70%), Gaps = 2/122 (1%)
Frame = +1
Query: 10 QTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSFLRFDSVDH 69
QT+V+L+I KHLFSKE EKN+VFSP SL +LS++ GS GST +L FLRF S DH
Sbjct: 34 QTKVSLTIVKHLFSKES--EKNIVFSPLSLQVVLSIITAGSEGSTQQQLLDFLRFKSTDH 207
Query: 70 LNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTTHYKATLALVDFR 127
LN+ S LS +L D T S LSFV+ +W +K+L+L SFKQ+++ YKATL+ VDF+
Sbjct: 208 LNSSVSHQLSIILKDATSSGGPHLSFVDGVWLEKTLSLQSSFKQIVSEDYKATLSEVDFK 387
Query: 128 TK 129
K
Sbjct: 388 NK 393
>BF006197 weakly similar to GP|1310677|emb| protein z-type serpin {Hordeum
vulgare subsp. vulgare}, partial (7%)
Length = 468
Score = 76.6 bits (187), Expect = 2e-14
Identities = 39/87 (44%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Frame = +3
Query: 45 VMAVGSAGSTLDELFSFLRFDSVDHLNTFFSQVLSTVLSDTTPS--FLLSFVNEMWADKS 102
++ G T +L FLR +DHLN+F S ++S +LSD PS LS +W D++
Sbjct: 3 IVTAACEGRTQQQLLEFLRSKFIDHLNSFTSHLVSIILSDAAPSGGSRLSLTQRVWVDQT 182
Query: 103 LALSHSFKQLMTTHYKATLALVDFRTK 129
L+L SFK+ M T YKATLA VDF+ K
Sbjct: 183 LSLQPSFKETMVTDYKATLASVDFQNK 263
>AW560154
Length = 470
Score = 67.8 bits (164), Expect = 7e-12
Identities = 34/64 (53%), Positives = 44/64 (68%), Gaps = 2/64 (3%)
Frame = +1
Query: 68 DHLNTFFSQVLSTVLSDTTPS--FLLSFVNEMWADKSLALSHSFKQLMTTHYKATLALVD 125
DHLN+F S ++S +LSD PS LSF +W D++L+L SFK+ M T YKATLA VD
Sbjct: 1 DHLNSFTSDLVSIILSDAAPSGGSRLSFTQRVWVDQTLSLQPSFKETMVTDYKATLASVD 180
Query: 126 FRTK 129
F+ K
Sbjct: 181 FQNK 192
>TC87143 similar to GP|17976705|emb|CAC80839. dihydrofolate synthetase
/folylpolyglutamate synthetase {Arabidopsis thaliana},
partial (41%)
Length = 920
Score = 36.6 bits (83), Expect = 0.018
Identities = 25/84 (29%), Positives = 38/84 (44%)
Frame = +2
Query: 264 GKLPRQKVRVRKFRIPKFKICFELEASNVLKELGVVSPFSKSNANFTKMVVSPLDELCVE 323
G++ +K + K RIP F + E +VLKE S N V SPLD +
Sbjct: 569 GEIAGEKAGILKHRIPTFTVTQPDEPMDVLKE-------KASQVNVPLQVTSPLDAKLLN 727
Query: 324 SIHHKASIEVNEEGTEAGVACSMC 347
+ K +E + AG+A ++C
Sbjct: 728 GL--KLGLEGEHQYVNAGLAVALC 793
>TC87864 similar to GP|16604637|gb|AAL24111.1 unknown protein {Arabidopsis
thaliana}, partial (42%)
Length = 1950
Score = 29.6 bits (65), Expect = 2.2
Identities = 25/82 (30%), Positives = 42/82 (50%), Gaps = 2/82 (2%)
Frame = +1
Query: 53 STLDELFSFLRFDSVDHLNTFFSQVLSTVL--SDTTPSFLLSFVNEMWADKSLALSHSFK 110
S +DE ++ RFDS+ H+ + + VL +D S L + +++ D LA +F
Sbjct: 223 SRIDETWTVARFDSLPHVVHILTSIYRPVLQFTDKVASMLPTKYSQLSNDGLLAFVENF- 399
Query: 111 QLMTTHYKATLALVDFRTKGDQ 132
+ H+ T+ VD+R KG Q
Sbjct: 400 --VKDHFLPTM-FVDYR-KGVQ 453
>AW774550 homologue to PIR|D84431|D84 probable endosomal protein [imported] -
Arabidopsis thaliana, partial (28%)
Length = 701
Score = 29.6 bits (65), Expect = 2.2
Identities = 21/88 (23%), Positives = 39/88 (43%), Gaps = 12/88 (13%)
Frame = -3
Query: 276 FRIPKFKICFELEASN------------VLKELGVVSPFSKSNANFTKMVVSPLDELCVE 323
FR PKFK F + +L +GV P+++ A FT +++ D++C+
Sbjct: 510 FRFPKFKSIFAAALGSGTQLFTLTIFIFMLALVGVFYPYNRG-ALFTACIITTQDQICLV 334
Query: 324 SIHHKASIEVNEEGTEAGVACSMCDVKL 351
++S+ A +CS+ V +
Sbjct: 333 LCKLRSSLATWPASAMASFSCSVA*VSV 250
>CB894765
Length = 679
Score = 28.9 bits (63), Expect = 3.7
Identities = 12/39 (30%), Positives = 18/39 (45%)
Frame = -3
Query: 310 TKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCD 348
+K V +D+L E +HH S T + C +CD
Sbjct: 263 SKCRVKMIDKLLREQLHHNTSFTCRVPQTRTNMNCKLCD 147
>CA858703 weakly similar to GP|22296462|db contains EST C25306(S5880)~similar
to Arabidopsis thaliana chromosome 2 At2g47990~unknown
protein, partial (21%)
Length = 406
Score = 28.5 bits (62), Expect = 4.8
Identities = 14/33 (42%), Positives = 18/33 (54%)
Frame = -2
Query: 60 SFLRFDSVDHLNTFFSQVLSTVLSDTTPSFLLS 92
SF + S HLN FF S++ + T P FL S
Sbjct: 264 SFSKSKSETHLNNFFLDTNSSITATTCPGFLPS 166
>AW208021 similar to SP|O70422|TFH4_ TFIIH basal transcription factor complex
p52 subunit (Basic transcription factor 52 kDa subunit),
partial (5%)
Length = 399
Score = 27.7 bits (60), Expect = 8.2
Identities = 15/43 (34%), Positives = 23/43 (52%)
Frame = +1
Query: 21 LFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSFLR 63
LFS+ EYQ NL+ + ++ + G T D++ SFLR
Sbjct: 112 LFSRVEYQLPNLIVGAITKESLYNAF---DNGITADQIVSFLR 231
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,358,171
Number of Sequences: 36976
Number of extensions: 169305
Number of successful extensions: 763
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 749
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 753
length of query: 402
length of database: 9,014,727
effective HSP length: 98
effective length of query: 304
effective length of database: 5,391,079
effective search space: 1638888016
effective search space used: 1638888016
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0175.7


