

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0175.11
(923 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
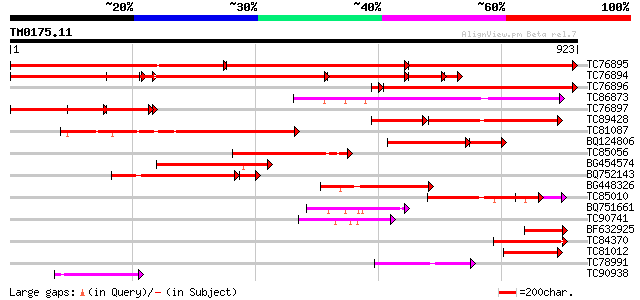
Score E
Sequences producing significant alignments: (bits) Value
TC76895 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protea... 555 0.0
TC76894 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protea... 535 0.0
TC76896 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protea... 588 e-172
TC86873 homologue to GP|530207|gb|AAA66338.1|| heat shock protei... 360 2e-99
TC76897 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protea... 338 6e-93
TC89428 homologue to GP|9651530|gb|AAF91178.1| ClpB {Phaseolus l... 231 1e-90
TC81087 homologue to GP|530207|gb|AAA66338.1|| heat shock protei... 290 2e-78
BQ124806 similar to SP|P42762|ERD1 ERD1 protein chloroplast pre... 213 2e-68
TC85056 homologue to GP|9651530|gb|AAF91178.1| ClpB {Phaseolus l... 203 2e-52
BG454574 similar to SP|P42762|ERD1 ERD1 protein chloroplast pre... 187 2e-47
BQ752143 similar to PIR|T39572|T39 probable proteinase subunit -... 147 1e-44
BG448326 similar to PIR|T51523|T51 clpB heat shock protein-like ... 167 2e-41
TC85010 weakly similar to PIR|T40514|T40514 Chaperonin hsp78p - ... 146 3e-35
BQ751661 weakly similar to PIR|T39572|T39 probable proteinase su... 97 2e-20
TC90741 similar to PIR|T51523|T51523 clpB heat shock protein-lik... 96 6e-20
BF632925 similar to GP|15425870|gb| heat shock protein 78 {Candi... 80 4e-15
TC84370 weakly similar to PIR|T39572|T39572 probable proteinase ... 80 4e-15
TC81012 similar to PIR|T51523|T51523 clpB heat shock protein-lik... 78 1e-14
TC78991 similar to GP|9759268|dbj|BAB09589.1 101 kDa heat shock ... 70 4e-12
TC90938 similar to GP|21592785|gb|AAM64734.1 unknown {Arabidopsi... 69 6e-12
>TC76895 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protease
ATP-binding subunit clpA homolog chloroplast precursor.
[Garden pea], complete
Length = 3425
Score = 555 bits (1430), Expect(3) = 0.0
Identities = 291/357 (81%), Positives = 315/357 (87%), Gaps = 1/357 (0%)
Frame = +2
Query: 1 MSRVLAQSITIPGLVCGRSHGHNNRSTMSRRSLKMMSTLQAPALRMSGFSGLRTYNNLDT 60
M+RVLAQSI +PGLV G + S S+RS+KMM + LRM GFSGLRT N+LDT
Sbjct: 230 MARVLAQSINVPGLVVGHKQSQHKGSGKSKRSVKMMCASRTIGLRMPGFSGLRTINHLDT 409
Query: 61 MLRPGLDFRSKVFGVTTSRKARASRCIPKAMFERFTEKAIKVIMLSQEEARRLGHNFVGT 120
MLRPGLDF +KV +SR+ARA R IP+AMFERFTEKAIKVIML+QEEARRLGHNFVGT
Sbjct: 410 MLRPGLDFHAKVSMAISSRRARAKRIIPRAMFERFTEKAIKVIMLAQEEARRLGHNFVGT 589
Query: 121 EQILLGLVGEGTGIAARVLKAMGISLKDARVEVEKTIGRGSGFVAVEIPFTPRAKRVLEL 180
EQILLGL+GEGTGIAA+VLK+MGI+LKDARVEVEK IGRGSGFVAVEIPFTPRAKRVLEL
Sbjct: 590 EQILLGLIGEGTGIAAKVLKSMGINLKDARVEVEKIIGRGSGFVAVEIPFTPRAKRVLEL 769
Query: 181 SLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQDLGADPNNIRAQVIRMVGES-NNET 239
SLEEARQLGHNYIGSEHLLLGLLREGEGVAARVL++LGADP NIR QVIRMVGES +N T
Sbjct: 770 SLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLENLGADPTNIRTQVIRMVGESADNVT 949
Query: 240 AGVAVGRGGSSNKMPTLEEYGTNLTKLANEGKLDPVVGRQQQIERVIQILGRRTKNNPCL 299
A VG G S+NK PTLEEYGTNLTKLA EGKLDPVVGRQ QIERV QILGRRTKNNPCL
Sbjct: 950 A--TVGSGSSNNKTPTLEEYGTNLTKLAEEGKLDPVVGRQPQIERVTQILGRRTKNNPCL 1123
Query: 300 VGEPGVGKTAIAEGLAQRIATGDVPETIEGKEVITLDMGLLVAGTKYRGEFEERLKK 356
+GEPGVGKTAIAEGLAQRIA GDVPETIEGK+VITLDMGLLVAGTKYRGEFEER ++
Sbjct: 1124IGEPGVGKTAIAEGLAQRIANGDVPETIEGKKVITLDMGLLVAGTKYRGEFEERFEE 1294
Score = 531 bits (1369), Expect(3) = 0.0
Identities = 267/297 (89%), Positives = 289/297 (96%)
Frame = +3
Query: 354 LKKLMEEIKQNDNIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEY 413
LKKLMEEIKQ+D+IILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEY
Sbjct: 1287 LKKLMEEIKQSDDIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEY 1466
Query: 414 RKHIEKDPALERRFQPVRVPEPTVDESIQILRGLRERYERHHKLSYTDDALVAASQLSHQ 473
RKHIEKDPALERRFQPV+VPEPTVDE+IQIL+GLRERYE HHKL YTDDAL+AA+QLS+Q
Sbjct: 1467 RKHIEKDPALERRFQPVKVPEPTVDETIQILKGLRERYEIHHKLRYTDDALIAAAQLSYQ 1646
Query: 474 YISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELDKEVRQIVKEKDEAVRNQDFEKAG 533
YISDRFLPDKAIDLIDEAGSRVRL+HAQLPEEA+ELDKEVR+IVKEK+E VRNQDFEKAG
Sbjct: 1647 YISDRFLPDKAIDLIDEAGSRVRLQHAQLPEEAKELDKEVRKIVKEKEEFVRNQDFEKAG 1826
Query: 534 ELRDREMDLKTQISALIEKGKEMSKAESEADGAGPVVTEVDIQHIVASWTGVPVDKVSSD 593
ELRD+EMDL+ QISAL+EKGKEMSKAESEA GP+VTEVDIQHIV+SWTG+PVDKVS+D
Sbjct: 1827 ELRDKEMDLRAQISALVEKGKEMSKAESEAADEGPIVTEVDIQHIVSSWTGIPVDKVSAD 2006
Query: 594 ESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPIASFIFSGPTGVGK 650
ESDRLLKME+TLHKRVIGQDEAV+AI RAIRRARVGLKNPNRPIASFIFSGPTGVG+
Sbjct: 2007 ESDRLLKMEDTLHKRVIGQDEAVEAISRAIRRARVGLKNPNRPIASFIFSGPTGVGE 2177
Score = 502 bits (1293), Expect(3) = 0.0
Identities = 255/275 (92%), Positives = 265/275 (95%)
Frame = +1
Query: 649 GKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRR 708
GKSELAK LA+YYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRR
Sbjct: 2173 GKSELAKALAAYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRR 2352
Query: 709 PYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGG 768
PYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGG
Sbjct: 2353 PYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGG 2532
Query: 769 RKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIM 828
R+IGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIM
Sbjct: 2533 RRIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIM 2712
Query: 829 LKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEI 888
L+EVF RLK KEI+L VTERFRDRVV+EGY+PSYGARPLRRAIMRLLEDSMAEKMLA EI
Sbjct: 2713 LREVFQRLKNKEIELQVTERFRDRVVDEGYNPSYGARPLRRAIMRLLEDSMAEKMLAREI 2892
Query: 889 KEGDSVIIDADSDGKVIVLNGSSGAPESLPEALPV 923
KEGDSVI+D DS+G VIVLNG+SG ESLPEAL +
Sbjct: 2893 KEGDSVIVDVDSEGNVIVLNGTSGTQESLPEALAI 2997
>TC76894 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protease
ATP-binding subunit clpA homolog chloroplast precursor.
[Garden pea], partial (71%)
Length = 2413
Score = 535 bits (1379), Expect(4) = 0.0
Identities = 273/309 (88%), Positives = 290/309 (93%)
Frame = +2
Query: 212 RVLQDLGADPNNIRAQVIRMVGESNNETAGVAVGRGGSSNKMPTLEEYGTNLTKLANEGK 271
RVL++LGADP NIR QVIRMVGE ++ G VG G S+NKMPTLEEYGTNLTKLA EGK
Sbjct: 839 RVLENLGADPTNIRTQVIRMVGEGA-DSVGATVGSGSSNNKMPTLEEYGTNLTKLAEEGK 1015
Query: 272 LDPVVGRQQQIERVIQILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVPETIEGKE 331
LDPVVGRQ QIERV QILGRRTKNNPCL+GEPGVGKTAIAEGLAQRIA GDVPETIEGK+
Sbjct: 1016 LDPVVGRQPQIERVTQILGRRTKNNPCLIGEPGVGKTAIAEGLAQRIANGDVPETIEGKK 1195
Query: 332 VITLDMGLLVAGTKYRGEFEERLKKLMEEIKQNDNIILFIDEVHTLIGAGAAEGAIDAAN 391
VITLDMGLLVAGTKYRGEFEERLKKLMEEIKQ+D IILFIDEVHTLIGAGAAEGAIDAAN
Sbjct: 1196 VITLDMGLLVAGTKYRGEFEERLKKLMEEIKQSDEIILFIDEVHTLIGAGAAEGAIDAAN 1375
Query: 392 ILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVRVPEPTVDESIQILRGLRERY 451
ILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPV+VPEPTV E+IQIL+GLRERY
Sbjct: 1376 ILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVKVPEPTVPETIQILKGLRERY 1555
Query: 452 ERHHKLSYTDDALVAASQLSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELDK 511
E HHKL YTD+ALVAA++LSHQYISDRFLPDKAIDLIDEAGSRVRL+HAQLPEEAR L+K
Sbjct: 1556 EIHHKLRYTDEALVAAAELSHQYISDRFLPDKAIDLIDEAGSRVRLQHAQLPEEARGLEK 1735
Query: 512 EVRQIVKEK 520
EVRQIVKE+
Sbjct: 1736 EVRQIVKEE 1762
Score = 318 bits (814), Expect = 8e-87
Identities = 168/226 (74%), Positives = 188/226 (82%), Gaps = 4/226 (1%)
Frame = +1
Query: 1 MSRVLAQSITIPGLVCGRSHGHNNR-STMSRRSLKMMSTLQAPALRMSGFSGLRTYNNLD 59
MSR LAQSI +PGLV GR H +NN+ + SRRS++MM T + + R+S +SGLRT N+LD
Sbjct: 193 MSRALAQSINVPGLVAGRRHVNNNKGAARSRRSVRMMFTTRTASPRLSSYSGLRTLNSLD 372
Query: 60 TMLRPGLDFRSKVF---GVTTSRKARASRCIPKAMFERFTEKAIKVIMLSQEEARRLGHN 116
+MLRPG DF SKV G ++ R SRC+ KAMFERFTEKAIKVIML+QEEARRLGHN
Sbjct: 373 SMLRPGQDFHSKVLTQIGTNRAKGGRGSRCVTKAMFERFTEKAIKVIMLAQEEARRLGHN 552
Query: 117 FVGTEQILLGLVGEGTGIAARVLKAMGISLKDARVEVEKTIGRGSGFVAVEIPFTPRAKR 176
FVGTEQILLGL+GEGTGIAA+VLK+MGI+LKDARVEVEK IGRGSGFVAVEIPFTPRAKR
Sbjct: 553 FVGTEQILLGLIGEGTGIAAKVLKSMGINLKDARVEVEKIIGRGSGFVAVEIPFTPRAKR 732
Query: 177 VLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQDLGADPN 222
VLELSLEEARQLGHNYIGSEH LLGLLREGEGVAA G PN
Sbjct: 733 VLELSLEEARQLGHNYIGSEHSLLGLLREGEGVAAPCS*KFGCRPN 870
Score = 227 bits (579), Expect(4) = 0.0
Identities = 114/133 (85%), Positives = 124/133 (92%)
Frame = +3
Query: 518 KEKDEAVRNQDFEKAGELRDREMDLKTQISALIEKGKEMSKAESEADGAGPVVTEVDIQH 577
+ KDEAVRNQ+FEKAGELRD+EMDLKTQISALIEK KEM+KAESEA G +VTEVDIQH
Sbjct: 1755 RRKDEAVRNQEFEKAGELRDKEMDLKTQISALIEKNKEMNKAESEAGDVGALVTEVDIQH 1934
Query: 578 IVASWTGVPVDKVSSDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPI 637
IVASWTG+PVDKVS DESDRLLKME+TLHKR+IGQ EAV+AI RAIRRARVGLKNPNRPI
Sbjct: 1935 IVASWTGIPVDKVSVDESDRLLKMEDTLHKRIIGQHEAVEAISRAIRRARVGLKNPNRPI 2114
Query: 638 ASFIFSGPTGVGK 650
ASFIFSGPTGVG+
Sbjct: 2115 ASFIFSGPTGVGE 2153
Score = 127 bits (319), Expect(4) = 0.0
Identities = 62/63 (98%), Positives = 62/63 (98%)
Frame = +1
Query: 649 GKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRR 708
GKSELAK LASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRR
Sbjct: 2149 GKSELAKALASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRR 2328
Query: 709 PYT 711
PYT
Sbjct: 2329 PYT 2337
Score = 71.6 bits (174), Expect = 1e-12
Identities = 37/85 (43%), Positives = 59/85 (68%), Gaps = 2/85 (2%)
Frame = +1
Query: 158 GRGSGFV--AVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQ 215
GRGS V A+ FT +A +V+ L+ EEAR+LGHN++G+E +LLGL+ EG G+AA+VL+
Sbjct: 445 GRGSRCVTKAMFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIAAKVLK 624
Query: 216 DLGADPNNIRAQVIRMVGESNNETA 240
+G + + R +V +++G + A
Sbjct: 625 SMGINLKDARVEVEKIIGRGSGFVA 699
Score = 53.1 bits (126), Expect(4) = 0.0
Identities = 25/28 (89%), Positives = 25/28 (89%)
Frame = +2
Query: 709 PYTVVLFDEIEKAHPDVFNMMLQILEDG 736
P VVLFDEI KAHPDVFNMMLQILEDG
Sbjct: 2330 PTRVVLFDEIXKAHPDVFNMMLQILEDG 2413
>TC76896 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protease
ATP-binding subunit clpA homolog chloroplast precursor.
[Garden pea], partial (35%)
Length = 1348
Score = 588 bits (1515), Expect(2) = e-172
Identities = 300/315 (95%), Positives = 311/315 (98%)
Frame = +1
Query: 609 VIGQDEAVKAICRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKTLASYYFGSEEAM 668
VIGQ EAV+AI RAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKTLASYYFGSEEAM
Sbjct: 61 VIGQHEAVEAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKTLASYYFGSEEAM 240
Query: 669 IRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNM 728
IRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNM
Sbjct: 241 IRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNM 420
Query: 729 MLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIK 788
MLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGR+IGFDLDYDEKDSSYNRIK
Sbjct: 421 MLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRRIGFDLDYDEKDSSYNRIK 600
Query: 789 SLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTER 848
SLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVF+RLKTKEI+LSVTER
Sbjct: 601 SLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFERLKTKEIELSVTER 780
Query: 849 FRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVIVLN 908
FR+RVV+EGY+PSYGARPLRRAIMRLLEDSMAEKMLA EIKEGDSVI+DADSDG VIVLN
Sbjct: 781 FRERVVDEGYNPSYGARPLRRAIMRLLEDSMAEKMLAREIKEGDSVIVDADSDGNVIVLN 960
Query: 909 GSSGAPESLPEALPV 923
GS+GAP+SLP+ALPV
Sbjct: 961 GSTGAPDSLPDALPV 1005
Score = 38.1 bits (87), Expect(2) = e-172
Identities = 18/19 (94%), Positives = 18/19 (94%)
Frame = +3
Query: 590 VSSDESDRLLKMEETLHKR 608
VS DESDRLLKMEETLHKR
Sbjct: 3 VSVDESDRLLKMEETLHKR 59
>TC86873 homologue to GP|530207|gb|AAA66338.1|| heat shock protein {Glycine
max}, partial (53%)
Length = 1475
Score = 360 bits (923), Expect = 2e-99
Identities = 201/498 (40%), Positives = 300/498 (59%), Gaps = 58/498 (11%)
Frame = +2
Query: 463 ALVAASQLSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELDK-------EVRQ 515
A+V A+QLS +YI+ R LPDKAIDL+DEA + VR++ PEE L++ E+
Sbjct: 2 AIVVAAQLSSRYITGRHLPDKAIDLVDEACANVRVQLDSQPEEIDNLERKRMQLEVELHA 181
Query: 516 IVKEKDEAVRNQDFE---KAGELRDREMDLKT----------QISALIEKGKEMSKAESE 562
+ KEKD+A + + + + +LRD+ LK +I L +K +E+ A E
Sbjct: 182 LEKEKDKASKARLVDVRRELDDLRDKLQPLKMKYSKEKERIDEIRRLKQKREELLFALQE 361
Query: 563 ADGAGPVVTEVDIQH--------------------------------------IVASWTG 584
A+ + D+++ +V+ WTG
Sbjct: 362 AERRYDLARAADLRYGAIEEVETAIKNLEGSTDGNTDENLMLTETVGPDQIAEVVSRWTG 541
Query: 585 VPVDKVSSDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPIASFIFSG 644
+PV ++ +E RL+ + + LH RV+GQD+AV A+ A+ R+R GL P +P SF+F G
Sbjct: 542 IPVTRLGQNEKARLVGLGDRLHTRVVGQDQAVNAVAEAVLRSRAGLGRPQQPTGSFLFLG 721
Query: 645 PTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEA 704
PTGVGK+ELAK LA F E ++R+DMSE+ME+H+VS+LIG+PPGYVG+ EGGQLTEA
Sbjct: 722 PTGVGKTELAKALAEQLFDDENQLVRIDMSEYMEQHSVSRLIGAPPGYVGHEEGGQLTEA 901
Query: 705 VRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVI 764
VRRRPY+VVLFDE+EKAH VFN +LQ+L+DGRLTD +GRTVDF+NT++IMTSN+G+ +
Sbjct: 902 VRRRPYSVVLFDEVEKAHTSVFNTLLQVLDDGRLTDGQGRTVDFRNTVIIMTSNLGAEHL 1081
Query: 765 EKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEI 824
G + + V +E++++FRPE LNRLDE++VF L+ +++++
Sbjct: 1082LSG----------LSGKCTMQAARDRVMQEVRRHFRPELLNRLDEVVVFDPLSHEQLRKV 1231
Query: 825 ADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKML 884
A + +K+V RL + I L+VT+ D ++ E YDP YGARP+R+ + + + ++ ++
Sbjct: 1232ARLQMKDVASRLAERGIALAVTDAALDYILAESYDPVYGARPIRKWLEKKVVTELSRMLI 1411
Query: 885 AGEIKEGDSVIIDADSDG 902
EI E + IDA G
Sbjct: 1412REEIDENTTGYIDAGPKG 1465
>TC76897 homologue to SP|P35100|CLPA_PEA ATP-dependent clp protease
ATP-binding subunit clpA homolog chloroplast precursor.
[Garden pea], partial (18%)
Length = 826
Score = 338 bits (867), Expect = 6e-93
Identities = 180/239 (75%), Positives = 199/239 (82%), Gaps = 5/239 (2%)
Frame = +2
Query: 1 MSRVLAQSITIPGLVCGRSH-GHNNRSTMSR-RSLKMMSTLQAPALRMSGFSGLRTYNNL 58
MSRVLAQSI +PGLV GR H +NN S SR RS++MM Q +R+SGFSGLR N+L
Sbjct: 89 MSRVLAQSINVPGLVAGRRHVNNNNGSVRSRPRSVRMMYATQTATMRLSGFSGLRPLNSL 268
Query: 59 DTMLRPGLDFRSKVF---GVTTSRKARASRCIPKAMFERFTEKAIKVIMLSQEEARRLGH 115
D+M RP DF SKV G +R R SRC+ +AMFERFTEKAIKVIML+QEEARRLGH
Sbjct: 269 DSMFRPRQDFHSKVLTQIGTNRARGGRGSRCVTRAMFERFTEKAIKVIMLAQEEARRLGH 448
Query: 116 NFVGTEQILLGLVGEGTGIAARVLKAMGISLKDARVEVEKTIGRGSGFVAVEIPFTPRAK 175
NFVGTEQILLGL+GEGTGI A+VLK+MGI+LKD+RVEVEK IGRGSGFVAVEIPFTPRAK
Sbjct: 449 NFVGTEQILLGLIGEGTGIGAKVLKSMGINLKDSRVEVEKIIGRGSGFVAVEIPFTPRAK 628
Query: 176 RVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQDLGADPNNIRAQVIRMVGE 234
RVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVL++LGADP NI VI M GE
Sbjct: 629 RVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLENLGADPTNIXTXVIXMXGE 805
Score = 69.7 bits (169), Expect = 5e-12
Identities = 36/85 (42%), Positives = 58/85 (67%), Gaps = 2/85 (2%)
Frame = +2
Query: 158 GRGSGFV--AVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQ 215
GRGS V A+ FT +A +V+ L+ EEAR+LGHN++G+E +LLGL+ EG G+ A+VL+
Sbjct: 344 GRGSRCVTRAMFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIGAKVLK 523
Query: 216 DLGADPNNIRAQVIRMVGESNNETA 240
+G + + R +V +++G + A
Sbjct: 524 SMGINLKDSRVEVEKIIGRGSGFVA 598
Score = 65.1 bits (157), Expect = 1e-10
Identities = 32/67 (47%), Positives = 46/67 (67%)
Frame = +2
Query: 95 FTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVLKAMGISLKDARVEVE 154
FT +A +V+ LS EEAR+LGHN++G+E +LLGL+ EG G+AARVL+ +G + V
Sbjct: 611 FTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLENLGADPTNIXTXVI 790
Query: 155 KTIGRGS 161
G G+
Sbjct: 791 XMXGEGA 811
>TC89428 homologue to GP|9651530|gb|AAF91178.1| ClpB {Phaseolus lunatus},
partial (32%)
Length = 1227
Score = 231 bits (590), Expect(2) = 1e-90
Identities = 117/218 (53%), Positives = 160/218 (72%)
Frame = +2
Query: 682 VSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDS 741
VS+L+G+PPGYVGY EGGQLTE VRRRPY+VVLFDEIEKAH DVFN++LQ+L+DGR+TDS
Sbjct: 317 VSRLVGAPPGYVGYEEGGQLTEVVRRRPYSVVLFDEIEKAHHDVFNILLQLLDDGRITDS 496
Query: 742 KGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRP 801
+GRTV F N +LIMTSN+GS I + D+K + Y+++K V E +Q FRP
Sbjct: 497 QGRTVSFTNCVLIMTSNIGSHHILE-----TLSSTQDDKIAVYDQMKRQVVELARQTFRP 661
Query: 802 EFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPS 861
EF+NR+DE IVF+ L E+ +I ++ ++ V RLK K+IDL TE + G+DP+
Sbjct: 662 EFMNRIDEYIVFQPLDSSEISKIVELQMERVKGRLKQKKIDLHYTEEAVKLLGVLGFDPN 841
Query: 862 YGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDAD 899
+GARP++R I +L+E+ +A +L G+ KE DS+I+DAD
Sbjct: 842 FGARPVKRVIQQLVENEIAMGVLRGDFKEEDSIIVDAD 955
Score = 121 bits (303), Expect(2) = 1e-90
Identities = 56/92 (60%), Positives = 74/92 (79%)
Frame = +1
Query: 589 KVSSDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPIASFIFSGPTGV 648
K E ++L+ +E+ LHKRVIGQD AVK++ AIRR+R GL +PNRPIASF+F GPTGV
Sbjct: 37 KPPQTEREKLVFLEQVLHKRVIGQDIAVKSVADAIRRSRAGLSDPNRPIASFMFMGPTGV 216
Query: 649 GKSELAKTLASYYFGSEEAMIRLDMSEFMERH 680
GK+EL K LA+Y F +E A++R+DMSE+ME+H
Sbjct: 217 GKTELGKALANYLFNTENALVRIDMSEYMEKH 312
>TC81087 homologue to GP|530207|gb|AAA66338.1|| heat shock protein {Glycine
max}, partial (41%)
Length = 1318
Score = 290 bits (742), Expect = 2e-78
Identities = 164/399 (41%), Positives = 244/399 (61%), Gaps = 11/399 (2%)
Frame = +2
Query: 84 SRCIPKAMF----ERFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVL 139
S I K+ F E+FT K + + + E A GH + + LV + GI + +
Sbjct: 152 SHSINKSSFTMNPEKFTHKTNEALAGAHELAMTSGHAQITPLHLASILVSDPNGIFFQAI 331
Query: 140 KAMGISLKDARVEVEKTIGRGSGFVAV------EIPFTPRAKRVLELSLEEARQLGHNYI 193
++ +++ VE+ + + + E+P + + + + + G ++
Sbjct: 332 S--NVAGEESARAVERVLKQALKKLPSQSPPPDEVPGSTALIKAIRRAQAAQKSRGDTHL 505
Query: 194 GSEHLLLGLLREGEGVAARVLQDLGADPNNIRAQVIRMVGESNNETAGVAVGRGGSSNKM 253
+ L+LG+L + + + ++ G + ++ +V ++ G+ G V
Sbjct: 506 AVDQLILGILEDSQ--IGDLFKEAGVAVSRVKTEVEKLRGKD-----GKKVESASGDTNF 664
Query: 254 PTLEEYGTNLTKLANEGKLDPVVGRQQQIERVIQILGRRTKNNPCLVGEPGVGKTAIAEG 313
L+ YG +L + A GKLDPV+GR ++I RV++IL RRTKNNP L+GEPGVGKTA+ EG
Sbjct: 665 QALKTYGRDLVEQA--GKLDPVIGRDEEIRRVVRILSRRTKNNPVLIGEPGVGKTAVVEG 838
Query: 314 LAQRIATGDVPETIEGKEVITLDMGLLVAGTKYRGEFEERLKKLMEEIKQNDN-IILFID 372
LAQRI GDVP + +I LDMG LVAG KYRGEFEERLK +++E+++ + +ILFID
Sbjct: 839 LAQRIVRGDVPSNLADVRLIALDMGALVAGAKYRGEFEERLKAVLKEVEEAEGKVILFID 1018
Query: 373 EVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVRV 432
E+H ++GAG EG++DAAN+ KP LARG+L+CIGATTL+EYRK++EKD A ERRFQ V V
Sbjct: 1019EIHLVLGAGRTEGSMDAANLFKPMLARGQLRCIGATTLEEYRKYVEKDAAFERRFQQVYV 1198
Query: 433 PEPTVDESIQILRGLRERYERHHKLSYTDDALVAASQLS 471
EP+V ++I ILRGL+ERYE HH + D A+V A+QLS
Sbjct: 1199AEPSVPDTISILRGLKERYEGHHGVRIQDRAIVVAAQLS 1315
>BQ124806 similar to SP|P42762|ERD1 ERD1 protein chloroplast precursor.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (20%)
Length = 679
Score = 213 bits (541), Expect(2) = 2e-68
Identities = 100/136 (73%), Positives = 122/136 (89%)
Frame = +2
Query: 615 AVKAICRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMS 674
AV AI R ++R+RVGL++P RPIA+ +F GPTGVGK+ELAK+LA+ YFGSE MIRLDMS
Sbjct: 2 AVSAISRFVKRSRVGLQDPGRPIATLLFCGPTGVGKTELAKSLAACYFGSETNMIRLDMS 181
Query: 675 EFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILE 734
E+MERH+VSKL+GSPPGYVGY EGG LTEA+RR+P+TVVLFDEIEKAHPD+FN++LQ++E
Sbjct: 182 EYMERHSVSKLLGSPPGYVGYGEGGILTEAIRRKPFTVVLFDEIEKAHPDIFNILLQLME 361
Query: 735 DGRLTDSKGRTVDFKN 750
DG LTDS+GR V FKN
Sbjct: 362 DGHLTDSQGRRVSFKN 409
Score = 65.9 bits (159), Expect(2) = 2e-68
Identities = 34/61 (55%), Positives = 43/61 (69%)
Frame = +3
Query: 749 KNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLD 808
K L++MTSNVGSS I KG L D+K +SY+ +KS+V EEL+ YFRPE LNR+D
Sbjct: 405 KMHLVVMTSNVGSSAIAKGRHNSMGFLISDDKPTSYSGLKSMVIEELRTYFRPELLNRID 584
Query: 809 E 809
E
Sbjct: 585 E 587
>TC85056 homologue to GP|9651530|gb|AAF91178.1| ClpB {Phaseolus lunatus},
partial (25%)
Length = 743
Score = 203 bits (517), Expect = 2e-52
Identities = 103/195 (52%), Positives = 145/195 (73%)
Frame = +1
Query: 364 NDNIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYRKHIEKDPAL 423
N IILFIDE+HT++GAGA GA+DA N+LKP L RGEL+CIGATTL+EYRK+IEKDPAL
Sbjct: 13 NGQIILFIDEIHTVVGAGATSGAMDAGNLLKPMLGRGELRCIGATTLNEYRKYIEKDPAL 192
Query: 424 ERRFQPVRVPEPTVDESIQILRGLRERYERHHKLSYTDDALVAASQLSHQYISDRFLPDK 483
ERRFQ V +P+V+++I ILRGLRERYE HH + +D ALV+A+ L+ +YI++RFLPDK
Sbjct: 193 ERRFQQVFCCQPSVEDTISILRGLRERYELHHGVKISDSALVSAAVLADRYITERFLPDK 372
Query: 484 AIDLIDEAGSRVRLRHAQLPEEARELDKEVRQIVKEKDEAVRNQDFEKAGELRDREMDLK 543
AIDL+DEA +++++ P E E+D+ V ++ E ++ D +KA ++R L+
Sbjct: 373 AIDLVDEAAAKLKMEITSKPTELDEIDRAVLKL--EMEKLSLKSDTDKAS--KERLSKLE 540
Query: 544 TQISALIEKGKEMSK 558
+S L +K KE+++
Sbjct: 541 NDLSLLKQKQKELAE 585
>BG454574 similar to SP|P42762|ERD1 ERD1 protein chloroplast precursor.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (19%)
Length = 672
Score = 187 bits (475), Expect = 2e-47
Identities = 97/193 (50%), Positives = 134/193 (69%), Gaps = 5/193 (2%)
Frame = +1
Query: 240 AGVAVGRGGSSNKMPTLEEYGTNLTKLANEGKLDPVVGRQQQIERVIQILGRRTKNNPCL 299
AG A +K L ++ +LT A+ G +DPV+GR+ +++R+IQIL R+TK+NP L
Sbjct: 94 AGSAAKTKDKKDKKNALSQFCVDLTARASVGLIDPVIGREVEVQRIIQILCRKTKSNPIL 273
Query: 300 VGEPGVGKTAIAEGLAQRIATGDVPETIEGKEVITLDMGLLVAGTKYRGEFEERLKKLME 359
+GE GVGKTAIAEGLA I+ +V + K V++LD+GLL+AG K RGE E+R+ KL++
Sbjct: 274 LGEAGVGKTAIAEGLAILISRAEVAPFLLTKRVMSLDVGLLMAGAKERGELEDRVTKLIK 453
Query: 360 EIKQNDNIILFIDEVHTLI-----GAGAAEGAIDAANILKPALARGELQCIGATTLDEYR 414
+I ++ ++ILFIDEVHTL+ G G D AN+LKP+L RG+ QCI +TT+DEYR
Sbjct: 454 DIIESGDVILFIDEVHTLVQSGTTGRGNKGSGFDIANLLKPSLGRGQFQCIASTTIDEYR 633
Query: 415 KHIEKDPALERRF 427
H EKD AL RF
Sbjct: 634 LHFEKDKALAXRF 672
>BQ752143 similar to PIR|T39572|T39 probable proteinase subunit - fission
yeast (Schizosaccharomyces pombe), partial (18%)
Length = 774
Score = 147 bits (371), Expect(2) = 1e-44
Identities = 78/208 (37%), Positives = 126/208 (60%), Gaps = 1/208 (0%)
Frame = +2
Query: 167 EIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQDLGADPNNIRA 226
++ P VL + E + +I +HL+ L + +Q + N +A
Sbjct: 62 QVSVAPTFHAVLRKAQELQKTQKDTFIAVDHLIQALAEDN------TIQTCLREANVPKA 223
Query: 227 QVIRMVGESNNETAGVAVGRGGSSNKMPTLEEYGTNLTKLANEGKLDPVVGRQQQIERVI 286
+++ T V + + L ++ ++T LA + K+DPV+GR+++I RV+
Sbjct: 224 KLVHDAVSQIRGTKRVDSKNADTEEEHENLAKFTIDMTALARDKKIDPVIGREEEIRRVV 403
Query: 287 QILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVPETIEGKEVITLDMGLLVAGTKY 346
+IL RRTKNNP L+GEPGVGKT + EGLAQRI DVP+ ++ ++++LD+G LVAG+KY
Sbjct: 404 RILSRRTKNNPVLIGEPGVGKTTVVEGLAQRIVNRDVPDNLKACKLLSLDVGALVAGSKY 583
Query: 347 RGEFEERLKKLMEEIKQN-DNIILFIDE 373
RGEFEER+K +++EI+ + + I+LF+DE
Sbjct: 584 RGEFEERMKGVLKEIEDSKEMIVLFVDE 667
Score = 52.0 bits (123), Expect(2) = 1e-44
Identities = 24/36 (66%), Positives = 31/36 (85%), Gaps = 1/36 (2%)
Frame = +1
Query: 374 VHTLIGAGAA-EGAIDAANILKPALARGELQCIGAT 408
+H L+GAG++ EG +DAAN+LKP LARG+L CIGAT
Sbjct: 667 IHLLMGAGSSGEGGMDAANLLKPMLARGQLHCIGAT 774
>BG448326 similar to PIR|T51523|T51 clpB heat shock protein-like -
Arabidopsis thaliana, partial (22%)
Length = 646
Score = 167 bits (423), Expect = 2e-41
Identities = 87/192 (45%), Positives = 125/192 (64%), Gaps = 8/192 (4%)
Frame = +2
Query: 507 RELDKEVRQIVKEKDEAVRNQDFEKAGELR-------DREMD-LKTQISALIEKGKEMSK 558
+ + +E+ ++ E +A R D +A EL+ R+++ + ++ + GK M +
Sbjct: 98 QSIKEEIDRVNLEIQQAEREYDLNRAAELKYGSLNSLQRQLESAEKELDEYMNSGKSMLR 277
Query: 559 AESEADGAGPVVTEVDIQHIVASWTGVPVDKVSSDESDRLLKMEETLHKRVIGQDEAVKA 618
E VT DI IV+ WTG+PV K+ E ++LL +EE LHKRV+GQD AVKA
Sbjct: 278 EE---------VTGSDIAEIVSKWTGIPVSKLQQSEREKLLYLEEVLHKRVVGQDPAVKA 430
Query: 619 ICRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFME 678
+ AI+R+R GL +P+RPIASF+F GPTGVGK+ELA TLASY F +E A++ +DMSE ME
Sbjct: 431 VAEAIQRSRAGLSDPHRPIASFMFMGPTGVGKTELAXTLASYMFNTEXALVXIDMSESME 610
Query: 679 RHTVSKLIGSPP 690
+H S+LIG+PP
Sbjct: 611 KHAXSRLIGAPP 646
>TC85010 weakly similar to PIR|T40514|T40514 Chaperonin hsp78p - fission
yeast (Schizosaccharomyces pombe), partial (23%)
Length = 904
Score = 146 bits (369), Expect = 3e-35
Identities = 79/203 (38%), Positives = 124/203 (60%), Gaps = 13/203 (6%)
Frame = +3
Query: 680 HTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLT 739
HT+S+LIG+P GYVGY + GQLTEAVRR+PY V+LFDE EKAH D+ ++LQ+L++G LT
Sbjct: 9 HTISRLIGAPSGYVGYEDAGQLTEAVRRKPYAVLLFDEFEKAHRDISALLLQVLDEGYLT 188
Query: 740 DSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRI----KSLVTEEL 795
D++G VDF+NT++++TSN+G+ ++ +G D + K+++ I + V + +
Sbjct: 189 DAQGHKVDFRNTIIVLTSNLGADIL------VGADPLHPYKETADGEIDPDVRKAVMDVV 350
Query: 796 KQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKT---------KEIDLSVT 846
+ PEFLNR+D IVF++L +++I DI + V D + L+
Sbjct: 351 AANYAPEFLNRIDSFIVFKRLALEALRDIVDISSRRVADSPRR*THLP*RT*SRS*LACG 530
Query: 847 ERFRDRVVEEGYDPSYGARPLRR 869
R R +V E P++ R +R
Sbjct: 531 ARLRPQVRSEAAKPAHHQRDWKR 599
Score = 61.6 bits (148), Expect = 1e-09
Identities = 33/85 (38%), Positives = 48/85 (55%), Gaps = 2/85 (2%)
Frame = +1
Query: 824 IADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKM 883
++ L E+ RL K I L V RD + E GYDP +GARPL R I + + +A+K+
Sbjct: 436 LSTFRLGELQTRLDDKRISLDVPNHVRDWLAERGYDPKFGARPLNRLITNEIGNGLADKI 615
Query: 884 LAGEIKEGD--SVIIDADSDGKVIV 906
+ GE+K G+ +V I D G +V
Sbjct: 616 IRGELKMGETATVAIKEDKSGLEVV 690
>BQ751661 weakly similar to PIR|T39572|T39 probable proteinase subunit -
fission yeast (Schizosaccharomyces pombe), partial (22%)
Length = 708
Score = 97.4 bits (241), Expect = 2e-20
Identities = 74/234 (31%), Positives = 113/234 (47%), Gaps = 66/234 (28%)
Frame = +2
Query: 484 AIDLIDEAGSRVRLRHAQLPEEARELDKEVRQIV-------KEKDEAVRNQDFEKAGELR 536
AIDL+DEA + VR+ PE L++++RQI+ KEKDEA + + + + +
Sbjct: 5 AIDLVDEAAAAVRVARESQPEIIDSLERKLRQIMIEIAALEKEKDEASQTRLVQARKDAK 184
Query: 537 DREMDLKT--------------------------------------------QISALIEK 552
+ E +L+ Q A+ E+
Sbjct: 185 NVEEELEPLRQKYLSEIKRGEQIHLAKLKLDELEKRLEDAANAGDHAKAADLQYGAIPEQ 364
Query: 553 G---KEMSKAESEADGA--------GPVVTEV----DIQHIVASWTGVPVDKVSSDESDR 597
KE+ ++ AD A G +VT+V I IV+ WTG+PV ++ + E ++
Sbjct: 365 RAVIKELEAKKAAADAALNATGQDHGAMVTDVVTADHINEIVSRWTGIPVTRLKTTEKEK 544
Query: 598 LLKMEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPIASFIFSGPTGVGKS 651
L+ ME+ L K V+GQ EAV+A+ AIR R GL NPN+P SF+F GP+G GK+
Sbjct: 545 LIHMEKQLGKIVVGQREAVQAVSNAIRLQRSGLSNPNQP-PSFLFCGPSGTGKT 703
>TC90741 similar to PIR|T51523|T51523 clpB heat shock protein-like -
Arabidopsis thaliana, partial (24%)
Length = 710
Score = 95.9 bits (237), Expect = 6e-20
Identities = 67/217 (30%), Positives = 106/217 (47%), Gaps = 59/217 (27%)
Frame = +1
Query: 470 LSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELDKEVRQIVKEKDEAVRNQDF 529
LS +YIS RFLPDKAIDL+DEA +++++ P E+++ V ++ E+ + + D
Sbjct: 58 LSDRYISGRFLPDKAIDLVDEAAAKLKMEITSKPTALDEINRSVLKLEMERLSLMNDTDK 237
Query: 530 -----------------EKAGELRDR---EMDLKTQISALIEK----GKEMSKAESEAD- 564
EK GEL ++ E + T++ ++ E+ E+ +AE E D
Sbjct: 238 ASKDRLNRLETELSLLKEKQGELTEQWEHEKSVMTRLQSIKEEIDRVNLEIQQAEREYDL 417
Query: 565 -------------------------------GAGPVVTEV---DIQHIVASWTGVPVDKV 590
G + EV DI IV+ WTG+P+ K+
Sbjct: 418 NRAAELKYGSLNSLQRQLEGAEKELHEYMSSGKSMLREEVTGNDIGEIVSKWTGIPISKL 597
Query: 591 SSDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRAR 627
E ++LL +++ LHKRV+GQD AVKA+ AI+R+R
Sbjct: 598 QQSEREKLLYLQDELHKRVVGQDPAVKAVAEAIQRSR 708
>BF632925 similar to GP|15425870|gb| heat shock protein 78 {Candida
albicans}, partial (2%)
Length = 525
Score = 80.1 bits (196), Expect = 4e-15
Identities = 36/70 (51%), Positives = 53/70 (75%)
Frame = -3
Query: 839 KEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDA 898
+EID V+E +D V +EGYDP+YGA PLR+AI+ L+ + +AE +LA + KEGD+V ID
Sbjct: 520 EEIDFEVSESVKDLVCKEGYDPTYGASPLRKAIVNLIANPLAEALLAEKCKEGDTVFIDL 341
Query: 899 DSDGKVIVLN 908
D++G +V+N
Sbjct: 340 DANGNTLVIN 311
>TC84370 weakly similar to PIR|T39572|T39572 probable proteinase subunit -
fission yeast (Schizosaccharomyces pombe), partial (13%)
Length = 643
Score = 80.1 bits (196), Expect = 4e-15
Identities = 41/122 (33%), Positives = 77/122 (62%), Gaps = 2/122 (1%)
Frame = +3
Query: 788 KSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLK--TKEIDLSV 845
+ LV L+ PEFLNR++ ++F +LT+ E+++I DI L E+ RL+ +++ + V
Sbjct: 12 RELVMNALRNCSLPEFLNRINSTVIFNRLTRREIRKIVDIRLLEIQKRLEGNGRKVYIDV 191
Query: 846 TERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVI 905
+E +D + GY P+YGARPL R I + + + +A +L G +++G++ ++ +G++
Sbjct: 192 SEEAKDYLGNAGYSPAYGARPLSRLIEKEVLNKLAILILRGNVRDGENARVEL-INGRIT 368
Query: 906 VL 907
VL
Sbjct: 369 VL 374
>TC81012 similar to PIR|T51523|T51523 clpB heat shock protein-like -
Arabidopsis thaliana, partial (11%)
Length = 577
Score = 78.2 bits (191), Expect = 1e-14
Identities = 38/95 (40%), Positives = 62/95 (65%)
Frame = +3
Query: 805 NRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGA 864
NR+DE IVFR L + ++ I + L+ V R+ +++ + VTE + GYDPSYGA
Sbjct: 3 NRVDEYIVFRPLDRDQISSIVRLQLERVQKRVADRKMKIRVTEPAIQLLGSLGYDPSYGA 182
Query: 865 RPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDAD 899
RP++R I + +E+ +A+ +L GE KE D+++ID +
Sbjct: 183 RPVKRVIQQNVENELAKGILRGEFKEEDTILIDTE 287
>TC78991 similar to GP|9759268|dbj|BAB09589.1 101 kDa heat shock protein;
HSP101-like protein {Arabidopsis thaliana}, partial
(13%)
Length = 1370
Score = 70.1 bits (170), Expect = 4e-12
Identities = 43/166 (25%), Positives = 84/166 (49%), Gaps = 1/166 (0%)
Frame = +2
Query: 594 ESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGL-KNPNRPIASFIFSGPTGVGKSE 652
++D ++ +TL ++V Q +A AI A+ + ++G K ++ +F+GP +GK
Sbjct: 314 DTDSFKRLLKTLTEKVWWQQDAASAIATAVTQCKLGNGKRRSKGDTWLMFTGPDRIGKKR 493
Query: 653 LAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTV 712
+A L+ GS +I L + + G T ++ E +RR P++V
Sbjct: 494 MAAALSELVSGSNPIVISLAQRRGDGDSNAHQ-------FRGKTVLDRIVETIRRNPHSV 652
Query: 713 VLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 758
++ ++I++A+ + + + +E GR DS GR + N + I+TSN
Sbjct: 653 IMLEDIDEANTLLRGNIKRAMEQGRFPDSHGREISLGNVMFILTSN 790
>TC90938 similar to GP|21592785|gb|AAM64734.1 unknown {Arabidopsis
thaliana}, partial (53%)
Length = 691
Score = 69.3 bits (168), Expect = 6e-12
Identities = 44/149 (29%), Positives = 79/149 (52%), Gaps = 3/149 (2%)
Frame = +2
Query: 73 FGVTTSRKARASRCIPKAMFERFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGT 132
F + TS R S P +++ KAIK +S EAR+L GTE +++G++ EGT
Sbjct: 227 FSLPTSNPERVS---PGQEIPKWSSKAIKSFAMSAIEARKLKFTTTGTEALIMGILIEGT 397
Query: 133 GIAARVLKAMGISLKDARVEVEKTIGRGSGFVAV--EIPFTPRAKRVLELSLE-EARQLG 189
+A++ L+A GI+L R E+ K +G F P T A+R L+ +++ + + +
Sbjct: 398 NLASKFLRANGITLFKVRDEIVKILGEVDPFTETPERPPMTDDAQRALDWAVDRKLKSVD 577
Query: 190 HNYIGSEHLLLGLLREGEGVAARVLQDLG 218
+ + H++LG+ + + ++L LG
Sbjct: 578 GGEVTTAHIILGIWSDVDSPGHKILSKLG 664
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.136 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,285,697
Number of Sequences: 36976
Number of extensions: 206797
Number of successful extensions: 1194
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 1147
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1171
length of query: 923
length of database: 9,014,727
effective HSP length: 105
effective length of query: 818
effective length of database: 5,132,247
effective search space: 4198178046
effective search space used: 4198178046
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0175.11


