

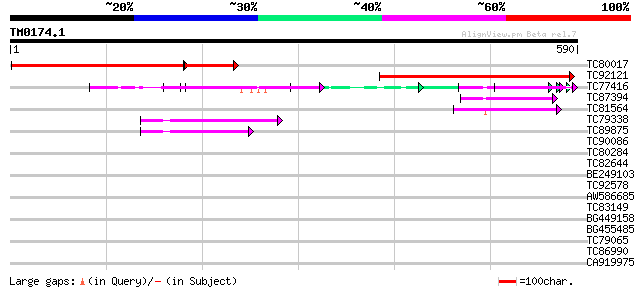
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0174.1
(590 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80017 similar to GP|10177301|dbj|BAB10562. gene_id:MDC12.17~un... 312 e-108
TC92121 weakly similar to GP|10177302|dbj|BAB10563. gene_id:MDC1... 332 2e-91
TC77416 similar to GP|18139887|gb|AAL60196.1 O-linked N-acetyl g... 74 2e-13
TC87394 similar to GP|3582779|gb|AAC69180.1| peroxisomal targeti... 51 1e-06
TC81564 weakly similar to PIR|E85082|E85082 hypothetical protein... 49 5e-06
TC79338 homologue to GP|20259245|gb|AAM14358.1 putative N-termin... 44 1e-04
TC89875 similar to GP|20259245|gb|AAM14358.1 putative N-terminal... 44 2e-04
TC90086 similar to PIR|G86185|G86185 hypothetical protein [impor... 38 0.013
TC80284 similar to PIR|T04740|T04740 hypothetical protein F6G17.... 37 0.028
TC82644 similar to GP|21537266|gb|AAM61607.1 unknown {Arabidopsi... 37 0.028
BE249103 weakly similar to GP|6630450|gb| F23N19.10 {Arabidopsis... 36 0.048
TC92578 similar to GP|10177072|dbj|BAB10514. contains similarity... 33 0.40
AW586685 similar to PIR|T01081|T01 hypothetical protein T10P11.3... 32 0.69
TC83149 similar to GP|13561982|gb|AAK30594.1 flagelliform silk p... 31 1.2
BG449158 similar to GP|17979432|gb putative TPR repeat nuclear p... 31 1.5
BG455485 31 1.5
TC79065 homologue to SP|P12204|YCF3_TOBAC Photosystem I assembly... 30 2.6
TC86990 similar to GP|22136840|gb|AAM91764.1 unknown protein {Ar... 29 4.5
CA919975 weakly similar to GP|3819697|emb| BnMAP4K alpha1 {Brass... 28 7.6
>TC80017 similar to GP|10177301|dbj|BAB10562. gene_id:MDC12.17~unknown
protein {Arabidopsis thaliana}, partial (45%)
Length = 879
Score = 312 bits (799), Expect(2) = e-108
Identities = 156/185 (84%), Positives = 170/185 (91%), Gaps = 1/185 (0%)
Frame = +2
Query: 3 RLTNDENSQDKSLLSKDTDSTEGEGKKSHKLGKCRSRPSKTDS-LDCGGDADVDQHVQGA 61
RL NDENSQDKSLLSKDT+S EGEGK +KLGKCRS+PSKTDS +DCG DAD DQHVQGA
Sbjct: 167 RLINDENSQDKSLLSKDTNSNEGEGKLLNKLGKCRSKPSKTDSSIDCGADADGDQHVQGA 346
Query: 62 PSSREEKVSSMKTGLIHVARKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPE 121
PS+REEKVSSMKTGL+HVARKMPKNAHAHFILGLM+QRLNQPQKAIL YEKAEEILLRPE
Sbjct: 347 PSAREEKVSSMKTGLVHVARKMPKNAHAHFILGLMYQRLNQPQKAILAYEKAEEILLRPE 526
Query: 122 TEIERPDLLSLVQIHHAQCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVW 181
EI+R + L+LVQIHHAQCLI+ESSSENSSD+ELEPHEL+EI+SKLKES Q DIRQAAVW
Sbjct: 527 VEIDRAEFLALVQIHHAQCLIIESSSENSSDQELEPHELEEIISKLKESTQSDIRQAAVW 706
Query: 182 NTLGF 186
N GF
Sbjct: 707 NYTGF 721
Score = 97.8 bits (242), Expect(2) = e-108
Identities = 50/56 (89%), Positives = 53/56 (94%)
Frame = +3
Query: 183 TLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQEL 238
TLGFILLKTGRVQSAISVLSSLLAI+PENYDCLGNLGIAYLQIG+LELSAK E+
Sbjct: 711 TLGFILLKTGRVQSAISVLSSLLAISPENYDCLGNLGIAYLQIGDLELSAKSVPEV 878
>TC92121 weakly similar to GP|10177302|dbj|BAB10563.
gene_id:MDC12.18~pir||F69210~similar to unknown protein
{Arabidopsis thaliana}, partial (76%)
Length = 845
Score = 332 bits (852), Expect = 2e-91
Identities = 165/203 (81%), Positives = 182/203 (89%)
Frame = +3
Query: 385 AGLAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQ 444
A L M HKAQHEIS+AYESEQ L E+EE AV SLKQA+AEDPDD V+WHQLG+HSLC +
Sbjct: 15 ARLQMSHKAQHEISAAYESEQDGLKEIEECAVSSLKQAIAEDPDDAVQWHQLGLHSLCAR 194
Query: 445 QFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILS 504
+FKTSQKYLKAAVACD+GCSY WSNLGVSLQLSEE SQAE+ YK AL LATKQ+AHAILS
Sbjct: 195 EFKTSQKYLKAAVACDKGCSYAWSNLGVSLQLSEEQSQAEEVYKWALSLATKQEAHAILS 374
Query: 505 NLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSD 564
N+GI YR +KKY+ AKAMFTKSLELQPGYAPAFNNLGLVF+AEGLLEEAK+CFEKALQSD
Sbjct: 375 NMGILYRQQKKYELAKAMFTKSLELQPGYAPAFNNLGLVFIAEGLLEEAKHCFEKALQSD 554
Query: 565 PLLDAAKSNLVKVVTMSKICKGL 587
+LDAAKSNL+KV TMSKICK L
Sbjct: 555 SMLDAAKSNLIKVATMSKICKDL 623
Score = 30.8 bits (68), Expect = 1.5
Identities = 14/64 (21%), Positives = 32/64 (49%)
Frame = +3
Query: 179 AVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQEL 238
A+ + +G + + + + A ++ + L + P NLG+ ++ G LE + CF++
Sbjct: 363 AILSNMGILYRQQKKYELAKAMFTKSLELQPGYAPAFNNLGLVFIAEGLLEEAKHCFEKA 542
Query: 239 ILKD 242
+ D
Sbjct: 543 LQSD 554
>TC77416 similar to GP|18139887|gb|AAL60196.1 O-linked N-acetyl glucosamine
transferase {Arabidopsis thaliana}, partial (94%)
Length = 3465
Score = 73.6 bits (179), Expect = 2e-13
Identities = 84/396 (21%), Positives = 143/396 (35%), Gaps = 13/396 (3%)
Frame = +2
Query: 184 LGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQ 243
LG I + ++ L I P +C GN+ A+ + GN++L+ + + I
Sbjct: 509 LGAIYYQLHDFDMCVAKNEEALRIEPHFAECYGNMANAWKEKGNIDLAIRYYLIAIELRP 688
Query: 244 NHPVALVNYAALLL-----------CKYASVVAG--AGASASEGALADQVMAANVAKECL 290
N A N A+ + C+ A + A ++ G L A C
Sbjct: 689 NFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEAYSCY 868
Query: 291 LAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAE 350
L A++ A W NLA F SGD + + ++A KL+P+
Sbjct: 869 LEALRIQPTFAIAWSNLAGLFMESGDFNRALQYYKEAVKLKPS----------------- 997
Query: 351 RSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTE 410
P A+ L V+KA
Sbjct: 998 -----------------------------FPDAYLNLGNVYKA---------------LG 1045
Query: 411 MEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNL 470
M + A+ + A+ P+ + + L Q + + K A+ACD ++NL
Sbjct: 1046MPQEAIACYQHALQTRPNYGMAYGNLASIHYEQGQLDMAILHYKQAIACDPRFLEAYNNL 1225
Query: 471 GVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQ 530
G +L+ +A + Y Q L L L+NLG Y A + + +L +
Sbjct: 1226GNALKDVGRVEEAIQCYNQCLSLQPNHPQ--ALTNLGNIYMEWNMVAAAASYYKATLNVT 1399
Query: 531 PGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPL 566
G + +NNL +++ +G +A C+ + L+ DPL
Sbjct: 1400TGLSAPYNNLAIIYKQQGNYADAISCYNEVLRIDPL 1507
Score = 53.5 bits (127), Expect = 2e-07
Identities = 62/282 (21%), Positives = 107/282 (36%)
Frame = +2
Query: 293 AIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERS 352
A++ + A +GN+A A+ G+ + + A +L PN +A+ M++ +
Sbjct: 569 ALRIEPHFAECYGNMANAWKEKGNIDLAIRYYLIAIELRPNFADAWSNLASAYMRKGRLT 748
Query: 353 QDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEME 412
E A R + + A + L + KAQ + AY
Sbjct: 749 ------------EAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEAYS---------- 862
Query: 413 ERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGV 472
C L +A+ P + W L + + F + +Y K AV + NLG
Sbjct: 863 ----CYL-EALRIQPTFAIAWSNLAGLFMESGDFNRALQYYKEAVKLKPSFPDAYLNLGN 1027
Query: 473 SLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPG 532
+ P +A Y+ AL T+ NL + + + A + +++ P
Sbjct: 1028VYKALGMPQEAIACYQHAL--QTRPNYGMAYGNLASIHYEQGQLDMAILHYKQAIACDPR 1201
Query: 533 YAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
+ A+NNLG G +EEA C+ + L P A +NL
Sbjct: 1202FLEAYNNLGNALKDVGRVEEAIQCYNQCLSLQPNHPQALTNL 1327
Score = 52.4 bits (124), Expect = 5e-07
Identities = 35/111 (31%), Positives = 57/111 (50%), Gaps = 1/111 (0%)
Frame = +2
Query: 468 SNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAIL-SNLGIFYRHEKKYQRAKAMFTKS 526
SNLG ++ +A Y +AL + Q AI SNL + + RA + ++
Sbjct: 809 SNLGNLMKAQGLVQEAYSCYLEALRI---QPTFAIAWSNLAGLFMESGDFNRALQYYKEA 979
Query: 527 LELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKV 577
++L+P + A+ NLG V+ A G+ +EA C++ ALQ+ P A NL +
Sbjct: 980 VKLKPSFPDAYLNLGNVYKALGMPQEAIACYQHALQTRPNYGMAYGNLASI 1132
Score = 51.2 bits (121), Expect = 1e-06
Identities = 61/258 (23%), Positives = 105/258 (40%), Gaps = 14/258 (5%)
Frame = +2
Query: 84 PKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSLVQIHHAQCLIL 143
P A+ LG +++ L PQ+AI Y+ A + RP + +L S IH+ Q
Sbjct: 992 PSFPDAYLNLGNVYKALGMPQEAIACYQHA--LQTRPNYGMAYGNLAS---IHYEQ---- 1144
Query: 144 ESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSS 203
+L + K+++ D R +N LG L GRV+ AI +
Sbjct: 1145 --------------GQLDMAILHYKQAIACDPRFLEAYNNLGNALKDVGRVEEAIQCYNQ 1282
Query: 204 LLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELI-----LKDQNHPVALV-----NYA 253
L++ P + L NLG Y++ + +A ++ + L + +A++ NYA
Sbjct: 1283 CLSLQPNHPQALTNLGNIYMEWNMVAAAASYYKATLNVTTGLSAPYNNLAIIYKQQGNYA 1462
Query: 254 ALLLCKYASVV----AGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAY 309
+ C Y V+ A + G ++ + A + + AI A NLA
Sbjct: 1463 DAISC-YNEVLRIDPLAADGLVNRGNTYKEIGRVSDAIQDYIRAITVRPTMAEAHANLAS 1639
Query: 310 AFSISGDHRSSSKCLEKA 327
A+ SG ++ K +A
Sbjct: 1640 AYKDSGHVEAAVKSYRQA 1693
Score = 47.0 bits (110), Expect = 2e-05
Identities = 81/407 (19%), Positives = 138/407 (33%)
Frame = +2
Query: 178 AAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQE 237
A W+ L ++ GR+ A LAI P D NLG G ++ + C+ E
Sbjct: 695 ADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEAYSCYLE 874
Query: 238 LILKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKAD 297
+ +A N A L + D A KE A+K
Sbjct: 875 ALRIQPTFAIAWSNLAGLFM-----------------ESGDFNRALQYYKE----AVKLK 991
Query: 298 VKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSE 357
+ NL + G + + C + A + PN Y +A + Q
Sbjct: 992 PSFPDAYLNLGNVYKALGMPQEAIACYQHALQTRPN-----YGMAYGNLASIHYEQGQL- 1153
Query: 358 LLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVC 417
D +++ A A +A + + +A + V E A+
Sbjct: 1154 ----------------DMAILHYKQAIACDPRFLEAYNNLGNALKDVGRV-----EEAIQ 1270
Query: 418 SLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLS 477
Q ++ P+ P LG + + Y KA + G S ++NL + +
Sbjct: 1271 CYNQCLSLQPNHPQALTNLGNIYMEWNMVAAAASYYKATLNVTTGLSAPYNNLAIIYKQQ 1450
Query: 478 EEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAF 537
+ A Y + L + A L N G Y+ + A + +++ ++P A A
Sbjct: 1451 GNYADAISCYNEVLRI--DPLAADGLVNRGNTYKEIGRVSDAIQDYIRAITVRPTMAEAH 1624
Query: 538 NNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSKIC 584
NL + G +E A + +AL A NL+ T+ +C
Sbjct: 1625 ANLASAYKDSGHVEAAVKSYRQALILRTDFPEATCNLLH--TLQCVC 1759
Score = 43.5 bits (101), Expect = 2e-04
Identities = 56/270 (20%), Positives = 103/270 (37%)
Frame = +2
Query: 161 KEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGI 220
+E ++ + ++Q + L I + G++ AI +A P + NLG
Sbjct: 1052 QEAIACYQHALQTRPNYGMAYGNLASIHYEQGQLDMAILHYKQAIACDPRFLEAYNNLGN 1231
Query: 221 AYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQV 280
A +G +E + +C+ + + NHP AL N + + ++ V A
Sbjct: 1232 ALKDVGRVEEAIQCYNQCLSLQPNHPQALTNLGNIYM-EWNMVAA--------------- 1363
Query: 281 MAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYA 340
AA+ K A + + + NLA + G++ + C + +++P +
Sbjct: 1364 -AASYYK----ATLNVTTGLSAPYNNLAIIYKQQGNYADAISCYNEVLRIDP-LAADGLV 1525
Query: 341 VATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSA 400
+ KE R D I+D ++ PT + +A ++SA
Sbjct: 1526 NRGNTYKEIGRVSD--------------AIQDYIRAITVRPT-------MAEAHANLASA 1642
Query: 401 YESEQHVLTEMEERAVCSLKQAVAEDPDDP 430
Y+ HV E AV S +QA+ D P
Sbjct: 1643 YKDSGHV-----EAAVKSYRQALILRTDFP 1717
Score = 42.7 bits (99), Expect = 4e-04
Identities = 25/86 (29%), Positives = 47/86 (54%)
Frame = +2
Query: 505 NLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSD 564
N+ ++ + A + ++EL+P +A A++NL ++ +G L EA C +AL +
Sbjct: 608 NMANAWKEKGNIDLAIRYYLIAIELRPNFADAWSNLASAYMRKGRLTEAAQCCRQALAIN 787
Query: 565 PLLDAAKSNLVKVVTMSKICKGLLKE 590
PL+ A SNL ++ +GL++E
Sbjct: 788 PLMVDAHSNLGNLMK----AQGLVQE 853
Score = 38.9 bits (89), Expect = 0.006
Identities = 34/174 (19%), Positives = 65/174 (36%)
Frame = +2
Query: 160 LKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLG 219
++E S E+++ A W+ L + +++G A+ + + P D NLG
Sbjct: 845 VQEAYSCYLEALRIQPTFAIAWSNLAGLFMESGDFNRALQYYKEAVKLKPSFPDAYLNLG 1024
Query: 220 IAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQ 279
Y +G + + C+Q + N+ +A N A++ +G L
Sbjct: 1025NVYKALGMPQEAIACYQHALQTRPNYGMAYGNLASI--------------HYEQGQLDMA 1162
Query: 280 VMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPN 333
++ A C D + + NL A G + +C + L+PN
Sbjct: 1163ILHYKQAIAC-------DPRFLEAYNNLGNALKDVGRVEEAIQCYNQCLSLQPN 1303
>TC87394 similar to GP|3582779|gb|AAC69180.1| peroxisomal targeting sequence
1 receptor {Nicotiana tabacum}, partial (65%)
Length = 1656
Score = 50.8 bits (120), Expect = 1e-06
Identities = 31/101 (30%), Positives = 57/101 (55%)
Frame = +3
Query: 470 LGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLEL 529
LGV LS E +A A++QAL L K Q +++ + LG + + A A + ++L+L
Sbjct: 792 LGVLYNLSREYDKAIAAFEQALKL--KPQDYSLWNKLGATQANSVQSADAIAAYQQALDL 965
Query: 530 QPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAA 570
+P Y A+ N+G+ + +G+ +E+ + +AL +P + A
Sbjct: 966 KPNYVRAWANMGISYANQGMYDESIRYYVRALAMNPKAENA 1088
Score = 33.9 bits (76), Expect = 0.18
Identities = 42/235 (17%), Positives = 99/235 (41%), Gaps = 9/235 (3%)
Frame = +3
Query: 273 EGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEP 332
+G L++ V+A L A + + +++ W L A + + D + + + +A + +P
Sbjct: 414 KGLLSEAVLA-------LEAEVLKNPENSEGWRLLGIAHAENDDDQQAIAAMMRAQEADP 572
Query: 333 NCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTA--WAGLAMV 390
+ A+ E E++ L + N + G + E+ + +A +A +
Sbjct: 573 TNLEVLLALGVSHTNELEQNAALKYLFGWLRNHP----KYGTIAPPEMSDSLYYADVARL 740
Query: 391 HKAQHEISSAYESEQHV-------LTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCT 443
+ + S +++ H+ L+ ++A+ + +QA+ P D W++LG +
Sbjct: 741 FN-EAAVISPDDADVHIVLGVLYNLSREYDKAIAAFEQALKLKPQDYSLWNKLGATQANS 917
Query: 444 QQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQ 498
Q + + A+ W+N+G+S ++ + Y +AL + K +
Sbjct: 918 VQSADAIAAYQQALDLKPNYVRAWANMGISYANQGMYDESIRYYVRALAMNPKAE 1082
Score = 30.4 bits (67), Expect = 2.0
Identities = 43/234 (18%), Positives = 90/234 (38%), Gaps = 50/234 (21%)
Frame = +3
Query: 52 ADVDQHVQGAPSSREEKVSSMKTGLIHVA--------RKMPKNAHAHFILGLMHQRLNQP 103
+D++ +V G P+ +E + GL+ A K P+N+ +LG+ H +
Sbjct: 351 SDLNPYV-GHPNPLKEGQDLFRKGLLSEAVLALEAEVLKNPENSEGWRLLGIAHAENDDD 527
Query: 104 QKAILVYEKAEE-------ILLR----PETEIERPDLLSLV------------------- 133
Q+AI +A+E +LL E+E+ L +
Sbjct: 528 QQAIAAMMRAQEADPTNLEVLLALGVSHTNELEQNAALKYLFGWLRNHPKYGTIAPPEMS 707
Query: 134 -QIHHAQCLILESSSENSSDKELEPH-----------ELKEILSKLKESVQFDIRQAAVW 181
+++A L + + S + + H E + ++ +++++ + ++W
Sbjct: 708 DSLYYADVARLFNEAAVISPDDADVHIVLGVLYNLSREYDKAIAAFEQALKLKPQDYSLW 887
Query: 182 NTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCF 235
N LG + + AI+ L + P N+GI+Y G + S + +
Sbjct: 888 NKLGATQANSVQSADAIAAYQQALDLKPNYVRAWANMGISYANQGMYDESIRYY 1049
>TC81564 weakly similar to PIR|E85082|E85082 hypothetical protein AT4g08320
[imported] - Arabidopsis thaliana, partial (46%)
Length = 1215
Score = 48.9 bits (115), Expect = 5e-06
Identities = 35/120 (29%), Positives = 56/120 (46%), Gaps = 7/120 (5%)
Frame = +3
Query: 462 GCS-YTWSNLGVSLQLSEEPSQAEKAYKQALLL-----ATKQQAHAILSNLGIFYRHEKK 515
GC + NL SL+ + K Y A+ L A +++ N Y +
Sbjct: 516 GCQQFNLKNLAESLKTLGNKAMQSKQYFDAIELYNCAIAIYEKSAVYYCNRAAAYTQINR 695
Query: 516 YQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEA-KYCFEKALQSDPLLDAAKSNL 574
Y A +S+E+ P Y+ A++ LGL + A+G +A F+KALQ DP ++ K N+
Sbjct: 696 YTEAIQDSLRSIEIDPNYSKAYSRLGLAYYAQGNYRDAIDKGFKKALQLDPNNESVKENI 875
>TC79338 homologue to GP|20259245|gb|AAM14358.1 putative N-terminal
acetyltransferase {Arabidopsis thaliana}, partial (24%)
Length = 904
Score = 44.3 bits (103), Expect = 1e-04
Identities = 32/148 (21%), Positives = 63/148 (41%)
Frame = +3
Query: 137 HAQCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQS 196
H + L ++ + N D++ E +EL +++ ++ D++ W+ G + +
Sbjct: 378 HGETLSMKGLTLNCMDRKSEAYEL------VRQGLKNDLKSHVCWHVYGLLYRSDREYRE 539
Query: 197 AISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALL 256
AI + L I P+N + L +L + Q+ +L + Q+L+ NH + + +A
Sbjct: 540 AIKCYRNALRIDPDNIEILRDLSLLQAQMRDLSGFVETRQQLLTLKSNHRMNWIGFAVSH 719
Query: 257 LCKYASVVAGAGASASEGALADQVMAAN 284
+ A A EG L D N
Sbjct: 720 HLNSNASKAIEILEAYEGTLEDDYPPEN 803
>TC89875 similar to GP|20259245|gb|AAM14358.1 putative N-terminal
acetyltransferase {Arabidopsis thaliana}, partial (38%)
Length = 1170
Score = 43.9 bits (102), Expect = 2e-04
Identities = 25/117 (21%), Positives = 55/117 (46%)
Frame = +3
Query: 137 HAQCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQS 196
H + L ++ + N D++ E +EL +++ ++ D++ W+ G + +
Sbjct: 228 HGETLSMKGLTLNCMDRKSEAYEL------VRQGLKNDLKSHVCWHVFGLLYRSDREYRE 389
Query: 197 AISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYA 253
AI + L I PEN + L +L + Q+ +L + Q+L+ NH + + ++
Sbjct: 390 AIKCYRNALRIDPENIEILRDLSLLQAQMRDLSGFVETRQQLLTLKPNHRMNWIGFS 560
>TC90086 similar to PIR|G86185|G86185 hypothetical protein [imported] -
Arabidopsis thaliana, partial (34%)
Length = 894
Score = 37.7 bits (86), Expect = 0.013
Identities = 31/117 (26%), Positives = 55/117 (46%)
Frame = +2
Query: 446 FKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSN 505
+K + K L+ A+ + +L +L E +A + +++A+ L K L N
Sbjct: 53 YKAAVKALEEAIFMKPDYADAHCDLASALHAMREDERAIEVFQKAIDL--KPGHIDALYN 226
Query: 506 LGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQ 562
L Y ++QRA M+T+ L + P + A N + + G EEAK ++AL+
Sbjct: 227 LSGLYMDLGRFQRASEMYTRVLAVWPNHWRAQLNKAVSLLGAGENEEAKKALKEALK 397
>TC80284 similar to PIR|T04740|T04740 hypothetical protein F6G17.110 -
Arabidopsis thaliana, partial (31%)
Length = 1645
Score = 36.6 bits (83), Expect = 0.028
Identities = 90/394 (22%), Positives = 151/394 (37%), Gaps = 16/394 (4%)
Frame = +2
Query: 188 LLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPV 247
L T AI +L SL++ + D + N Y Q LEL ++ Q +P+
Sbjct: 74 LCSTKDWSKAIRILDSLISQSTAIQD-ICNRAFCYSQ---LELHKHVIKDCDRAIQLNPL 241
Query: 248 ALVNYAALLLCKYASVVAGAGASA----SEGALADQVMAANVAK----ECLLAAIKADVK 299
L Y +L +A G A A +G Q +A++ + E LL K +
Sbjct: 242 LLQAY---ILKGHAFSALGRKADALLVWEQGYEQAQHHSADLKQLIELEELLVKAKQAIN 412
Query: 300 SAHIWGNLAY--AFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSE 357
S++ L+ A S S +R+ ++ E AKL N + +K A++ +E
Sbjct: 413 SSNETNGLSIPQAKSDSSSNRNLTETCESQAKLSGNTSDKSEVL----LKSADKFDARNE 580
Query: 358 LLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVC 417
L S GG D + P ++ +++ S ES + VLT E +
Sbjct: 581 LNSEGGESSKC-----DGQVNGSPD------IIDNLRYDSSDTSESCEKVLTNSGESSDS 727
Query: 418 SLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLS 477
+ + P + S + + + S+K+ A V+ + S V +LS
Sbjct: 728 NDAAEILRKPS-----FKFTFPSEKSSEARKSKKFSVARVSKTKSIS-------VDFRLS 871
Query: 478 EEPSQA-EKAYKQAL-----LLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQP 531
++ E Y A+ +L L G Y +++ A A FTK+++ P
Sbjct: 872 RGIAEVNEGKYAHAISIFDQILKEDSAYPEALIGRGTAYAFKRELHSAIADFTKAIQYNP 1051
Query: 532 GYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDP 565
A+ G A G EA KAL+ +P
Sbjct: 1052AAGEAWKRRGQARAALGEFVEAIEDLTKALEFEP 1153
>TC82644 similar to GP|21537266|gb|AAM61607.1 unknown {Arabidopsis
thaliana}, partial (70%)
Length = 937
Score = 36.6 bits (83), Expect = 0.028
Identities = 32/124 (25%), Positives = 55/124 (43%), Gaps = 1/124 (0%)
Frame = +2
Query: 468 SNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAIL-SNLGIFYRHEKKYQRAKAMFTKS 526
+ L V + E +Q E A + A + + + +I +N G+ + KY+ TK+
Sbjct: 374 NKLFVDAKYEEALTQYELALEVAPDMPSSVEIRSICHANRGVCFLKMGKYENTVKECTKA 553
Query: 527 LELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSKICKG 586
LEL P Y A G EEA +K L+ DP D A + ++ ++ + +
Sbjct: 554 LELNPMYVKALVRRGEAHEKLEHFEEAIADMKKILEIDPSNDQAGKAIRRLEPLAAVKRE 733
Query: 587 LLKE 590
+KE
Sbjct: 734 KMKE 745
>BE249103 weakly similar to GP|6630450|gb| F23N19.10 {Arabidopsis thaliana},
partial (20%)
Length = 613
Score = 35.8 bits (81), Expect = 0.048
Identities = 23/82 (28%), Positives = 38/82 (46%)
Frame = +1
Query: 493 LATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEE 552
+A H + SN K+Y+ A K +EL+P + + LG G ++
Sbjct: 124 IAVDATNHVLYSNRSAAQASLKRYKEALEDAQKVVELKPDWPKGHSRLGTAKQGLGDWDD 303
Query: 553 AKYCFEKALQSDPLLDAAKSNL 574
A +++ALQ +P +AAK L
Sbjct: 304 AIDAYKRALQLEPTNEAAKKAL 369
>TC92578 similar to GP|10177072|dbj|BAB10514. contains similarity to unknown
protein~dbj|BAA91048.1~gene_id:MKP11.12 {Arabidopsis
thaliana}, partial (36%)
Length = 1126
Score = 32.7 bits (73), Expect = 0.40
Identities = 43/228 (18%), Positives = 82/228 (35%), Gaps = 17/228 (7%)
Frame = +3
Query: 293 AIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPN----------------CMS 336
AIK + + +W NL + +S+ ++ + + K PN C
Sbjct: 411 AIK-EFEDLELWDNLIHCYSLLEKKATAVELIRKRLSERPNDPRLWCSLGDITNNDACYE 587
Query: 337 TRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVEL-PTAWAGLAMVHKAQH 395
V+ +R A+RS S + G E + ++ + S+ + P W
Sbjct: 588 KALEVSNNRSARAKRSLARS-AYNRGEYETSKVLWESAMSMNSMFPDGWFAFGAAALKAR 764
Query: 396 EISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKA 455
++ E+A+ + +AV DPD+ W+ + L ++ K + K
Sbjct: 765 DV---------------EKALDAFTRAVQLDPDNGEAWNNIACLHLIKKKSKEAFIAFKE 899
Query: 456 AVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAIL 503
A+ R W N SQA +A + L + ++ +L
Sbjct: 900 ALKFKRNSWQLWENYSHVAVDVGNISQALEAAQMVLDITKNKRVDTVL 1043
Score = 30.4 bits (67), Expect = 2.0
Identities = 32/155 (20%), Positives = 66/155 (41%)
Frame = +3
Query: 408 LTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTW 467
L E + AV +++ ++E P+DP W LG + Y KA + +
Sbjct: 468 LLEKKATAVELIRKRLSERPNDPRLWCSLGDIT------NNDACYEKALEVSNNRSARAK 629
Query: 468 SNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSL 527
+L S E ++ ++ A+ + + G + ++A FT+++
Sbjct: 630 RSLARSAYNRGEYETSKVLWESAMSMNSMFPDGWFA--FGAAALKARDVEKALDAFTRAV 803
Query: 528 ELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQ 562
+L P A+NN+ + + + +EA F++AL+
Sbjct: 804 QLDPDNGEAWNNIACLHLIKKKSKEAFIAFKEALK 908
>AW586685 similar to PIR|T01081|T01 hypothetical protein T10P11.3.2 -
Arabidopsis thaliana, partial (22%)
Length = 651
Score = 32.0 bits (71), Expect = 0.69
Identities = 15/27 (55%), Positives = 19/27 (69%)
Frame = +2
Query: 536 AFNNLGLVFVAEGLLEEAKYCFEKALQ 562
A NNLG VFV G L++A C+ KAL+
Sbjct: 95 ALNNLGSVFVDHGKLDQAADCYIKALK 175
>TC83149 similar to GP|13561982|gb|AAK30594.1 flagelliform silk protein
{Argiope trifasciata}, partial (3%)
Length = 1177
Score = 31.2 bits (69), Expect = 1.2
Identities = 25/95 (26%), Positives = 43/95 (44%), Gaps = 5/95 (5%)
Frame = +2
Query: 473 SLQLSEEPSQAEKAYKQALLLATK----QQAHAI-LSNLGIFYRHEKKYQRAKAMFTKSL 527
SL+ + A K Y A+ L T+ +A+ LSN + K + A+A ++
Sbjct: 248 SLKSRGNAAMASKDYPSAIALYTEALSLNPGNAVYLSNRAAAHSAAKDHSSARADAEAAV 427
Query: 528 ELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQ 562
+ P Y A++ LGL A G + A + K ++
Sbjct: 428 AIDPAYTKAWSRLGLARFALGDAKGAMEAYGKGIE 532
>BG449158 similar to GP|17979432|gb putative TPR repeat nuclear
phosphoprotein {Arabidopsis thaliana}, partial (26%)
Length = 693
Score = 30.8 bits (68), Expect = 1.5
Identities = 17/69 (24%), Positives = 32/69 (45%)
Frame = +1
Query: 189 LKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVA 248
+K G +SA+S +L + P+N + L L Y+Q+G + + ++ D A
Sbjct: 1 IKLGDFRSALSNFEKVLEVYPDNCETLKALAYIYVQLGQTDKGHEFIRKATKIDPRDAQA 180
Query: 249 LVNYAALLL 257
+ LL+
Sbjct: 181 FLELGELLI 207
>BG455485
Length = 677
Score = 30.8 bits (68), Expect = 1.5
Identities = 19/81 (23%), Positives = 35/81 (42%), Gaps = 3/81 (3%)
Frame = +3
Query: 8 ENSQDKSLLSKDTDSTEGEGKKSHKLGKCRSRPSKTDSLDC---GGDADVDQHVQGAPSS 64
ENS++ + ++ G GK S+ + + G DV+ +V G +
Sbjct: 30 ENSEEIEIKHNSENNNNGTGKVDTSTSTSTSKNVEVNGNTAKSGNGSGDVNVNVNGKVNK 209
Query: 65 REEKVSSMKTGLIHVARKMPK 85
E ++ KTG++HV P+
Sbjct: 210 PSEGSATGKTGVVHVQELKPE 272
>TC79065 homologue to SP|P12204|YCF3_TOBAC Photosystem I assembly protein
ycf3. [Common tobacco] {Nicotiana tabacum}, partial (70%)
Length = 1301
Score = 30.0 bits (66), Expect = 2.6
Identities = 18/73 (24%), Positives = 37/73 (50%), Gaps = 1/73 (1%)
Frame = +3
Query: 472 VSLQLSEEPSQAEKAYKQALLLATKQQAHA-ILSNLGIFYRHEKKYQRAKAMFTKSLELQ 530
+S Q ++A + Y +A+ L + IL N+G+ + ++ +A + ++LE
Sbjct: 1053 MSAQSEGNYAEALQNYYEAMRLEIDPYDRSYILYNIGLIHTSNGEHTKALEYYFRALERN 1232
Query: 531 PGYAPAFNNLGLV 543
P AFNN+ ++
Sbjct: 1233 PFLPQAFNNMAVI 1271
>TC86990 similar to GP|22136840|gb|AAM91764.1 unknown protein {Arabidopsis
thaliana}, partial (64%)
Length = 2354
Score = 29.3 bits (64), Expect = 4.5
Identities = 14/38 (36%), Positives = 21/38 (54%)
Frame = +2
Query: 231 SAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAG 268
SA+ F+ I +N+PVAL+NY +CK + G
Sbjct: 788 SARLFKMRIKFVKNYPVALINYTPWKICKLELIKISQG 901
>CA919975 weakly similar to GP|3819697|emb| BnMAP4K alpha1 {Brassica napus},
partial (12%)
Length = 768
Score = 28.5 bits (62), Expect = 7.6
Identities = 29/119 (24%), Positives = 49/119 (40%), Gaps = 3/119 (2%)
Frame = -2
Query: 316 DHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDS 375
D S + AA L + + + K R + S++ ++M S D
Sbjct: 761 DRNSMPSLKDSAANLAEAKAAIQGGRKVNARKRHSRGKINSDIQESKRDQMTSST-DSSR 585
Query: 376 SLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVC---SLKQAVAEDPDDPV 431
S E A G++ H A + SA +L+ +V SLK+A+A+DP+ P+
Sbjct: 584 SYREYIDAQRGMSKSHYASDDEESA-----RILSSSAPLSVLLIPSLKEAIADDPEGPI 423
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.129 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,178,763
Number of Sequences: 36976
Number of extensions: 227847
Number of successful extensions: 1098
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 1053
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1089
length of query: 590
length of database: 9,014,727
effective HSP length: 101
effective length of query: 489
effective length of database: 5,280,151
effective search space: 2581993839
effective search space used: 2581993839
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0174.1


