

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
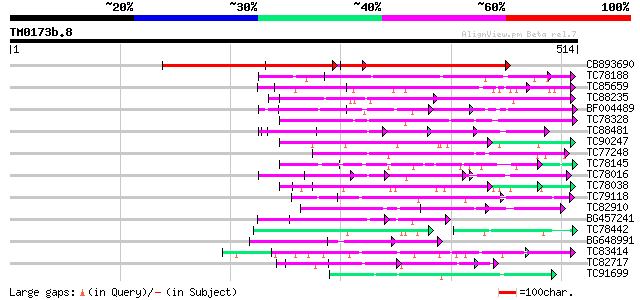
Query= TM0173b.8
(514 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB893690 weakly similar to GP|9759025|dbj gene_id:K6A12.9~simila... 266 e-140
TC78188 similar to PIR|C84870|C84870 probable splicing factor [i... 115 5e-26
TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein ... 105 4e-23
TC88235 similar to GP|12656803|gb|AAK00964.1 unknown protein {Or... 82 5e-16
BF004489 similar to GP|20161344|d contains ESTs AU086067(S4642) ... 78 9e-15
TC78328 similar to GP|11994333|dbj|BAB02292. WD-40 repeat protei... 74 1e-13
TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclea... 72 4e-13
TC90247 weakly similar to GP|21592898|gb|AAM64848.1 unknown {Ara... 71 9e-13
TC77248 similar to GP|13489182|gb|AAK27816.1 putative WD-repeat ... 66 4e-11
TC78145 similar to GP|15294240|gb|AAK95297.1 AT3g18860/MCB22_3 {... 66 4e-11
TC78016 similar to GP|21537191|gb|AAM61532.1 PRL1 protein {Arabi... 50 7e-11
TC78038 weakly similar to GP|21592898|gb|AAM64848.1 unknown {Ara... 62 5e-10
TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {... 61 9e-10
TC82910 similar to GP|19347844|gb|AAL86002.1 unknown protein {Ar... 60 2e-09
BG457241 homologue to PIR|F84600|F8 coatomer alpha subunit [impo... 60 3e-09
TC78442 homologue to GP|5733806|gb|AAD49742.1| G protein beta su... 51 3e-09
BG648991 similar to PIR|C86239|C86 protein T10O24.21 [imported] ... 59 3e-09
TC83414 weakly similar to GP|17381206|gb|AAL36415.1 unknown prot... 59 6e-09
TC82717 weakly similar to GP|21539583|gb|AAM53344.1 putative coa... 58 1e-08
TC91699 similar to GP|21592393|gb|AAM64344.1 unknown {Arabidopsi... 57 1e-08
>CB893690 weakly similar to GP|9759025|dbj gene_id:K6A12.9~similar to unknown
protein~sp|O15736 {Arabidopsis thaliana}, partial (59%)
Length = 916
Score = 266 bits (679), Expect(2) = e-140
Identities = 132/159 (83%), Positives = 146/159 (91%)
Frame = +2
Query: 139 QLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEANALYEEMVERLRASGLEQLARQQ 198
QLEQMT + +AEAENKML DR MLEKMKDAERLNEANALYE+MV+RLRASGLEQLAR+Q
Sbjct: 2 QLEQMTTKANEAEAENKMLTDRLMLEKMKDAERLNEANALYEDMVQRLRASGLEQLAREQ 181
Query: 199 VDGVVRRCEEGAEFFLDSNIPSTCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWD 258
VD +VRR EEGAEFF SNIPS CK+RLNAHEGGCAS+LFE+NSS+LITGGQD SVKVWD
Sbjct: 182 VDEIVRRSEEGAEFFSQSNIPSNCKYRLNAHEGGCASLLFEYNSSKLITGGQDHSVKVWD 361
Query: 259 TNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLY 297
TNTGS+ S+L GC+GSVLDLTIT+DNRSVIAASSSNNLY
Sbjct: 362 TNTGSLSSSLTGCLGSVLDLTITNDNRSVIAASSSNNLY 478
Score = 249 bits (637), Expect(2) = e-140
Identities = 123/154 (79%), Positives = 133/154 (85%)
Frame = +3
Query: 301 LNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCN 360
L + + +TLTGH DKVCAVD+SKVSSR VVSAAYD TIKVWDLMKGYCTNTI+F S CN
Sbjct: 453 LQAAQTTYTLTGHTDKVCAVDISKVSSRLVVSAAYDGTIKVWDLMKGYCTNTIMFPSICN 632
Query: 361 ALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDN 420
ALCFS DGQTI+SGHVDGNLRLWDIQSG+LLSEVA HS AVTSISLSRNGN+ LTSG DN
Sbjct: 633 ALCFSTDGQTIFSGHVDGNLRLWDIQSGRLLSEVATHSHAVTSISLSRNGNIALTSGGDN 812
Query: 421 VHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISP 454
+HNLFDV SLEVC + R T N VASNWSRSCI P
Sbjct: 813 LHNLFDV*SLEVCGTLRATVNIVASNWSRSCICP 914
Score = 44.3 bits (103), Expect = 1e-04
Identities = 23/92 (25%), Positives = 45/92 (48%)
Frame = +3
Query: 233 CASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASS 292
CA + + +S +++ D ++KVWD G C+N L + D +++ +
Sbjct: 504 CAVDISKVSSRLVVSAAYDGTIKVWDLMKG-YCTNTIMFPSICNALCFSTDGQTIFSGHV 680
Query: 293 SNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSK 324
NL +WD+ SGR+ + H V ++ +S+
Sbjct: 681 DGNLRLWDIQSGRLLSEVATHSHAVTSISLSR 776
>TC78188 similar to PIR|C84870|C84870 probable splicing factor [imported] -
Arabidopsis thaliana, partial (95%)
Length = 1436
Score = 115 bits (287), Expect = 5e-26
Identities = 77/274 (28%), Positives = 125/274 (45%), Gaps = 7/274 (2%)
Frame = +1
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSN---LHGCIGSVLDLTITH 282
L H+ ++ F S + +G D+ + +W N C N L G +VLDL T
Sbjct: 301 LTGHQSAVYTMKFNPTGSVVASGSHDKEIFLW--NVHGDCKNFMVLKGHKNAVLDLHWTS 474
Query: 283 DNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVW 342
D +I+AS L +WD +G+ + H V + ++ VVS + D T K+W
Sbjct: 475 DGTQIISASPDKTLRLWDTETGKQIKKMVEHLSYVNSCCPTRRGPPLVVSGSDDGTAKLW 654
Query: 343 DLMKGYCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVT 402
D+ + T A+ FS IY+G +D ++++WD++ G++ + H +T
Sbjct: 655 DMRQRGSIQTFPDKYQITAVSFSDASDKIYTGGIDNDVKIWDLRKGEVTMTLQGHQDMIT 834
Query: 403 SISLSRNGNVVLTSGRDNVHNLFDVRSL----EVCSSFRDTGNRVASNWSRSCISPDNSH 458
S+ LS +G+ +LT+G D ++D+R + N + SPD S
Sbjct: 835 SMQLSPDGSYLLTNGMDCKLCIWDMRPYAPQNRCVKILEGHQHNFEKNLLKCGWSPDGSK 1014
Query: 459 VAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCC 492
V AGS+D V+IW I+ L H SV C
Sbjct: 1015VTAGSSDRMVYIWDTTSRRILYKLPGHNGSVNEC 1116
Score = 75.9 bits (185), Expect = 4e-14
Identities = 61/241 (25%), Positives = 116/241 (47%), Gaps = 13/241 (5%)
Frame = +1
Query: 286 SVIAASSSNN-LYVWDLNSGRVRH-TLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWD 343
SV+A+ S + +++W+++ L GHK+ V + + ++ ++SA+ D+T+++WD
Sbjct: 352 SVVASGSHDKEIFLWNVHGDCKNFMVLKGHKNAVLDLHWTSDGTQ-IISASPDKTLRLWD 528
Query: 344 LMKGYCTNTII-FASNCNALCFSMDGQT-IYSGHVDGNLRLWDI-QSGKLLSEVAAHSLA 400
G ++ S N+ C + G + SG DG +LWD+ Q G + +
Sbjct: 529 TETGKQIKKMVEHLSYVNSCCPTRRGPPLVVSGSDDGTAKLWDMRQRGSI--QTFPDKYQ 702
Query: 401 VTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVA 460
+T++S S + + T G DN ++D+R EV + + + + S +SPD S++
Sbjct: 703 ITAVSFSDASDKIYTGGIDNDVKIWDLRKGEVTMTLQGHQDMITSMQ----LSPDGSYLL 870
Query: 461 AGSADGSVHIWSVH----KNEIVSTLKEHT----SSVLCCRWSGIGKPLASADKNGIVCI 512
D + IW + +N V L+ H ++L C WS G + + + +V I
Sbjct: 871 TNGMDCKLCIWDMRPYAPQNRCVKILEGHQHNFEKNLLKCGWSPDGSKVTAGSSDRMVYI 1050
Query: 513 W 513
W
Sbjct: 1051W 1053
Score = 33.9 bits (76), Expect = 0.15
Identities = 19/69 (27%), Positives = 36/69 (51%), Gaps = 1/69 (1%)
Frame = +1
Query: 241 NSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVI-AASSSNNLYVW 299
+ S++ G D+ V +WDT + + L G GSV + + H N ++ + SS +Y+
Sbjct: 1003 DGSKVTAGSSDRMVYIWDTTSRRILYKLPGHNGSVNE-CVFHPNEPIVGSCSSDKQIYLG 1179
Query: 300 DLNSGRVRH 308
++ S R+
Sbjct: 1180 EI*STAFRN 1206
>TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein beta
subunit-like protein. [Alfalfa] {Medicago sativa},
complete
Length = 1261
Score = 105 bits (262), Expect = 4e-23
Identities = 83/302 (27%), Positives = 139/302 (45%), Gaps = 29/302 (9%)
Frame = +2
Query: 241 NSSRLITGGQDQSVKVW-----DTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNN 295
NS ++T +D+S+ +W D G L G V D+ ++ D + ++ S
Sbjct: 140 NSDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDVVLSSDGQFALSGSWDGE 319
Query: 296 LYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIF 355
L +WDLN+G GH V +V S + +R +VSA+ DRTIK+W+ + G C TI
Sbjct: 320 LRLWDLNAGTSARRFVGHTKDVLSVAFS-IDNRQIVSASRDRTIKLWNTL-GECKYTIQD 493
Query: 356 AS------NCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRN 409
+C S TI S D +++W++ + KL + +A HS V ++++S +
Sbjct: 494 GDAHSDWVSCVRFSPSTLQPTIVSASWDRTVKVWNLTNCKLRNTLAGHSGYVNTVAVSPD 673
Query: 410 GNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVH 469
G++ + G+D V L+D+ + S D G+ + + C SP N + + + S+
Sbjct: 674 GSLCASGGKDGVILLWDLAEGKRLYSL-DAGSIIHA----LCFSP-NRYWLCAATESSIK 835
Query: 470 IWSVHKNEIVSTLKEHTSS---------------VLCC---RWSGIGKPLASADKNGIVC 511
IW + IV LK + V+ C WS G L S +G+V
Sbjct: 836 IWDLESKSIVEDLKVDLKTEADAAIGGDTTTKKKVIYCTSLNWSADGSTLFSGYTDGVVR 1015
Query: 512 IW 513
+W
Sbjct: 1016VW 1021
Score = 91.7 bits (226), Expect = 6e-19
Identities = 71/273 (26%), Positives = 121/273 (44%), Gaps = 24/273 (8%)
Frame = +2
Query: 225 RLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDN 284
RL H ++ + ++G D +++WD N G+ G VL + + DN
Sbjct: 233 RLTGHSHFVQDVVLSSDGQFALSGSWDGELRLWDLNAGTSARRFVGHTKDVLSVAFSIDN 412
Query: 285 RSVIAASSSNNLYVWDLNSGRVRHTL---TGHKDKVCAVDVSKVSSR-HVVSAAYDRTIK 340
R +++AS + +W+ G ++T+ H D V V S + + +VSA++DRT+K
Sbjct: 413 RQIVSASRDRTIKLWN-TLGECKYTIQDGDAHSDWVSCVRFSPSTLQPTIVSASWDRTVK 589
Query: 341 VWDLMKGYCTNTIIFASN-CNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSL 399
VW+L NT+ S N + S DG SG DG + LWD+ GK L + A S+
Sbjct: 590 VWNLTNCKLRNTLAGHSGYVNTVAVSPDGSLCASGGKDGVILLWDLAEGKRLYSLDAGSI 769
Query: 400 AVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFR-------------DTGNR---- 442
+ ++ S N L + ++ ++D+ S + + DT +
Sbjct: 770 -IHALCFSPN-RYWLCAATESSIKIWDLESKSIVEDLKVDLKTEADAAIGGDTTTKKKVI 943
Query: 443 --VASNWSRSCISPDNSHVAAGSADGSVHIWSV 473
+ NW S D S + +G DG V +W +
Sbjct: 944 YCTSLNW-----SADGSTLFSGYTDGVVRVWGI 1027
Score = 77.8 bits (190), Expect = 9e-15
Identities = 61/218 (27%), Positives = 104/218 (46%), Gaps = 10/218 (4%)
Frame = +2
Query: 306 VRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMK-----GYCTNTIIFASN-C 359
+R T+ H D V A+ +S +V+A+ D++I +W L K G + S+
Sbjct: 80 LRGTMRAHTDVVTAIATPIDNSDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFV 259
Query: 360 NALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRD 419
+ S DGQ SG DG LRLWD+ +G H+ V S++ S + ++++ RD
Sbjct: 260 QDVVLSSDGQFALSGSWDGELRLWDLNAGTSARRFVGHTKDVLSVAFSIDNRQIVSASRD 439
Query: 420 NVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCI--SPDNSH--VAAGSADGSVHIWSVHK 475
L++ +L C G+ S+W SC+ SP + + S D +V +W++
Sbjct: 440 RTIKLWN--TLGECKYTIQDGD-AHSDWV-SCVRFSPSTLQPTIVSASWDRTVKVWNLTN 607
Query: 476 NEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
++ +TL H+ V S G AS K+G++ +W
Sbjct: 608 CKLRNTLAGHSGYVNTVAVSPDGSLCASGGKDGVILLW 721
>TC88235 similar to GP|12656803|gb|AAK00964.1 unknown protein {Oryza
sativa}, partial (91%)
Length = 1080
Score = 82.0 bits (201), Expect = 5e-16
Identities = 66/279 (23%), Positives = 128/279 (45%), Gaps = 10/279 (3%)
Frame = +2
Query: 245 LITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSG 304
L T D +++ W+ +G + V L IT D R +AA+ + ++ ++D+NS
Sbjct: 233 LATASYDHTIRFWEAKSGRCYRTIQYPDSQVNRLEITPDKR-YLAAAGNPHIRLFDVNSN 409
Query: 305 RVRHTLT--GHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNAL 362
+ ++ GH V AV + + S + D T+K+WDL C + N +
Sbjct: 410 SPQPVMSYDGHTSNVMAVGF-QCDGNWMYSGSEDGTVKIWDLRAPGCQREYESRAAVNTV 586
Query: 363 CFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAH-SLAVTSISLSRNGNVVLTSGRDNV 421
+ + SG +GN+R+WD+ + E+ AV S+++ +G++V+ + +
Sbjct: 587 VLHPNQTELISGDQNGNIRVWDLTANSCSCELVPEVDTAVRSLTVMWDGSLVVAANNNGT 766
Query: 422 HNLFD-VRSLEVCSSFRDTGNRVASN-WSRSCI-SPD----NSHVAAGSADGSVHIWSVH 474
++ +R + ++F A N + C+ SP+ + ++A S+D +V IW+V
Sbjct: 767 CYVWRLLRGTQTMTNFEPLHKLQAHNGYILKCVLSPEFCDPHRYLATASSDHTVKIWNVD 946
Query: 475 KNEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
+ TL H + C +S G L +A + +W
Sbjct: 947 DFTLEKTLIGHQRRLWDCVFSVDGAYLITASSDSTARLW 1063
Score = 62.0 bits (149), Expect = 5e-10
Identities = 44/166 (26%), Positives = 75/166 (44%), Gaps = 12/166 (7%)
Frame = +2
Query: 235 SILFEFNSSRLITGGQDQSVKVWDTNTGSV-CSNLHGCIGSVLDLTITHDNRSVIAASSS 293
+++ N + LI+G Q+ +++VWD S C + +V LT+ D V+AA+++
Sbjct: 581 TVVLHPNQTELISGDQNGNIRVWDLTANSCSCELVPEVDTAVRSLTVMWDGSLVVAANNN 760
Query: 294 NNLYVWDLNSGRVR-------HTLTGHKDKVCAVDVSKV---SSRHVVSAAYDRTIKVWD 343
YVW L G H L H + +S R++ +A+ D T+K+W+
Sbjct: 761 GTCYVWRLLRGTQTMTNFEPLHKLQAHNGYILKCVLSPEFCDPHRYLATASSDHTVKIWN 940
Query: 344 LMKGYCTNTIIFASNCNALC-FSMDGQTIYSGHVDGNLRLWDIQSG 388
+ T+I C FS+DG + + D RLW +G
Sbjct: 941 VDDFTLEKTLIGHQRRLWDCVFSVDGAYLITASSDSTARLWSTTTG 1078
>BF004489 similar to GP|20161344|d contains ESTs AU086067(S4642)
D41819(S4642)~similar to Arabidopsis thaliana chromosome
1 At1g49040~, partial (16%)
Length = 605
Score = 77.8 bits (190), Expect = 9e-15
Identities = 52/211 (24%), Positives = 101/211 (47%), Gaps = 2/211 (0%)
Frame = +1
Query: 306 VRHTLTGHKDKVCAV--DVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALC 363
+R TL GH V A+ D+ KV VS + D+++ VWD + +
Sbjct: 1 LRATLKGHTRTVRAISSDMGKV-----VSGSDDQSVLVWDKQTTQLLEELKGHDGPVSCV 165
Query: 364 FSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHN 423
++ G+ + + DG +++WD+++ + ++ V S AV + N ++ +GRD V N
Sbjct: 166 RTLSGERVLTASHDGTVKMWDVRTDRCVATVGRCSSAVLCMEYDDNVGILAAAGRDVVAN 345
Query: 424 LFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLK 483
++D+R+ + +G+ + W RS + V GS D + IWSV + + L
Sbjct: 346 MWDIRASR--QMHKLSGH---TQWIRS-LRMVGDTVITGSDDWTARIWSVSRGTCDAVLA 507
Query: 484 EHTSSVLCCRWSGIGKPLASADKNGIVCIWK 514
H +LC +S + + + + +G++ W+
Sbjct: 508 CHAGPILCVEYSSLDRGIITGSTDGLLRFWE 600
Score = 64.3 bits (155), Expect = 1e-10
Identities = 42/162 (25%), Positives = 81/162 (49%), Gaps = 3/162 (1%)
Frame = +1
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNR 285
L H+G S + + R++T D +VK+WD T + + C +VL + DN
Sbjct: 133 LKGHDGP-VSCVRTLSGERVLTASHDGTVKMWDVRTDRCVATVGRCSSAVLCMEY-DDNV 306
Query: 286 SVIAASSSNNL-YVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDL 344
++AA+ + + +WD+ + R H L+GH + ++ ++ V++ + D T ++W +
Sbjct: 307 GILAAAGRDVVANMWDIRASRQMHKLSGHTQWIRSL---RMVGDTVITGSDDWTARIWSV 477
Query: 345 MKGYCTNTIIFASNCNALC--FSMDGQTIYSGHVDGNLRLWD 384
+G C + ++ LC +S + I +G DG LR W+
Sbjct: 478 SRGTC-DAVLACHAGPILCVEYSSLDRGIITGSTDGLLRFWE 600
Score = 52.0 bits (123), Expect = 5e-07
Identities = 39/178 (21%), Positives = 72/178 (39%)
Frame = +1
Query: 244 RLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNS 303
++++G DQSV VWD T + L G G V T V+ AS + +WD+ +
Sbjct: 61 KVVSGSDDQSVLVWDKQTTQLLEELKGHDGPV-SCVRTLSGERVLTASHDGTVKMWDVRT 237
Query: 304 GRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALC 363
R T+ V ++ + + +A D +WD+ + + +
Sbjct: 238 DRCVATVGRCSSAVLCMEYDD-NVGILAAAGRDVVANMWDIRASRQMHKLSGHTQW-IRS 411
Query: 364 FSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNV 421
M G T+ +G D R+W + G + +A H+ + + S ++T D +
Sbjct: 412 LRMVGDTVITGSDDWTARIWSVSRGTCDAVLACHAGPILCVEYSSLDRGIITGSTDGL 585
>TC78328 similar to GP|11994333|dbj|BAB02292. WD-40 repeat protein-like
{Arabidopsis thaliana}, partial (88%)
Length = 1516
Score = 74.3 bits (181), Expect = 1e-13
Identities = 65/274 (23%), Positives = 124/274 (44%), Gaps = 4/274 (1%)
Frame = +1
Query: 245 LITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSG 304
LI+ +D S + + TG G G+V + + AS+ + VWD +G
Sbjct: 361 LISASKDASPMLRNGETGDWIGTFEGHKGAVWSCCLDANALRAATASADFSTKVWDALTG 540
Query: 305 RVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFAS--NCNAL 362
H+ HK V A S+ + +++ ++ ++++DL + + S + ++
Sbjct: 541 DALHSFE-HKHIVRACAFSE-DTHLLLTGGAEKILRIYDLNRPDAPPKEVDKSPGSVRSV 714
Query: 363 CFSMDGQTIYSGHVD-GNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNV 421
+ QTI S D G +RLWD+++GK++ + S +TS +S++G + T+ V
Sbjct: 715 AWLHSDQTILSSCTDMGGVRLWDVRTGKIVQTLDTKS-PMTSAEVSQDGRYITTADGSTV 891
Query: 422 HNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPD-NSHVAAGSADGSVHIWSVHKNEIVS 480
+D + S+ + N + + P + AG D VH++ H + ++
Sbjct: 892 -KFWDANHYGLVKSYD-----MPCNVESASLEPKFGNKFVAGGEDLWVHVFDFHTGDEIA 1053
Query: 481 TLKEHTSSVLCCRWSGIGKPLASADKNGIVCIWK 514
K H V C R+S G+ AS ++G + IW+
Sbjct: 1054CNKGHHGPVHCVRFSPGGESYASGSEDGTIRIWQ 1155
>TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclear
ribonucleoprotein [imported] - Arabidopsis thaliana,
partial (60%)
Length = 1836
Score = 72.4 bits (176), Expect = 4e-13
Identities = 44/211 (20%), Positives = 89/211 (41%)
Frame = +1
Query: 279 TITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRT 338
+ + D + + S + +W + + + TL GH + V S V H+ +A+ DRT
Sbjct: 970 SFSRDGKGLATCSFTGATKLWSMPNVKKVSTLKGHTQRATDVAYSPVHKNHLATASADRT 1149
Query: 339 IKVWDLMKGYCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHS 398
K W+ + F G+ + + D RLWD+++ + L HS
Sbjct: 1150 AKYWNDQGALLGTFKGHLERLARIAFHPSGKYLGTASYDKTWRLWDVETEEELLLQEGHS 1329
Query: 399 LAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSH 458
+V + +G++ + G D + ++D+R+ + + SP+ H
Sbjct: 1330 RSVYGLDFHHDGSLAASCGLDALARVWDLRTGRSVLALEGHVKSILG----ISFSPNGYH 1497
Query: 459 VAAGSADGSVHIWSVHKNEIVSTLKEHTSSV 489
+A G D + IW + K + + T+ H++ +
Sbjct: 1498 LATGGEDNTCRIWDLRKKKSLYTIPAHSNLI 1590
Score = 67.8 bits (164), Expect = 1e-11
Identities = 49/194 (25%), Positives = 80/194 (40%), Gaps = 2/194 (1%)
Frame = +1
Query: 234 ASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSS 293
A I F + L T D++ ++WD T G SV L HD +
Sbjct: 1213 ARIAFHPSGKYLGTASYDKTWRLWDVETEEELLLQEGHSRSVYGLDFHHDGSLAASCGLD 1392
Query: 294 NNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTI 353
VWDL +GR L GH + + S + H+ + D T ++WDL K TI
Sbjct: 1393 ALARVWDLRTGRSVLALEGHVKSILGISFSP-NGYHLATGGEDNTCRIWDLRKKKSLYTI 1569
Query: 354 IFASN-CNALCFS-MDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGN 411
SN + + F +G + + D ++W + K + ++ H VTS+ + +G
Sbjct: 1570 PAHSNLISQVKFEPQEGYFLVTASYDMTAKVWSSRDFKPVKTLSGHEAKVTSLDVLGDGG 1749
Query: 412 VVLTSGRDNVHNLF 425
++T D L+
Sbjct: 1750 YIVTVSHDRTIKLW 1791
Score = 58.5 bits (140), Expect = 6e-09
Identities = 37/156 (23%), Positives = 63/156 (39%), Gaps = 1/156 (0%)
Frame = +1
Query: 229 HEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVI 288
H + F + S + G D +VWD TG L G + S+L ++ + + +
Sbjct: 1324 HSRSVYGLDFHHDGSLAASCGLDALARVWDLRTGRSVLALEGHVKSILGISFSPNGYHLA 1503
Query: 289 AASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGY 348
N +WDL + +T+ H + + V +V+A+YD T KVW
Sbjct: 1504 TGGEDNTCRIWDLRKKKSLYTIPAHSNLISQVKFEPQEGYFLVTASYDMTAKVWSSRDFK 1683
Query: 349 CTNTII-FASNCNALCFSMDGQTIYSGHVDGNLRLW 383
T+ + +L DG I + D ++LW
Sbjct: 1684 PVKTLSGHEAKVTSLDVLGDGGYIVTVSHDRTIKLW 1791
Score = 50.8 bits (120), Expect = 1e-06
Identities = 31/118 (26%), Positives = 52/118 (43%), Gaps = 1/118 (0%)
Frame = +1
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTI-THDN 284
L H I F N L TGG+D + ++WD + + + +
Sbjct: 1441 LEGHVKSILGISFSPNGYHLATGGEDNTCRIWDLRKKKSLYTIPAHSNLISQVKFEPQEG 1620
Query: 285 RSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVW 342
++ AS VW + TL+GH+ KV ++DV ++V+ ++DRTIK+W
Sbjct: 1621 YFLVTASYDMTAKVWSSRDFKPVKTLSGHEAKVTSLDVLG-DGGYIVTVSHDRTIKLW 1791
Score = 30.0 bits (66), Expect = 2.2
Identities = 20/57 (35%), Positives = 28/57 (49%), Gaps = 3/57 (5%)
Frame = +1
Query: 453 SPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGK---PLASADK 506
S D +A S G+ +WS+ + VSTLK HT +S + K ASAD+
Sbjct: 976 SRDGKGLATCSFTGATKLWSMPNVKKVSTLKGHTQRATDVAYSPVHKNHLATASADR 1146
>TC90247 weakly similar to GP|21592898|gb|AAM64848.1 unknown {Arabidopsis
thaliana}, partial (60%)
Length = 1041
Score = 71.2 bits (173), Expect = 9e-13
Identities = 47/215 (21%), Positives = 94/215 (42%), Gaps = 22/215 (10%)
Frame = +1
Query: 245 LITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNS- 303
L+ G + +W+ TG + H + +V L + D+ +I+ S + VW L S
Sbjct: 361 LVGGALSGDIYLWEVETGRLLKKWHAHLRAVTCLVFSDDDSLLISGSEDGYVRVWSLFSL 540
Query: 304 ----------GRVRHTLTGHKDKVCAVDVSKVSSRHV-VSAAYDRTIKVWDLMKGYCTNT 352
++ + H +V V + + VSA+ DRT KVW L +G +
Sbjct: 541 FDDLRSREEKNLYEYSFSEHTMRVTDVVIGNGGCNAIIVSASDDRTCKVWSLSRGTLLRS 720
Query: 353 IIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGK----------LLSEVAAHSLAVT 402
I+F S NA+ Y+G DG + + + + + ++ + HS AVT
Sbjct: 721 IVFPSVINAIVLDPAEHVFYAGSSDGKIFIAALNTERITTIDNYGMHIIGSFSNHSKAVT 900
Query: 403 SISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFR 437
++ S+ N++++ D + +++ ++ + F+
Sbjct: 901 CLAYSKGRNLLISGSEDGIVRVWNAKTHNIVRMFK 1005
Score = 53.5 bits (127), Expect = 2e-07
Identities = 67/300 (22%), Positives = 118/300 (39%), Gaps = 31/300 (10%)
Frame = +1
Query: 245 LITGGQDQSVKVWDTNTGSVCSNLHGCIG---SVLDLTITHDNRSVIAASSSNNLYVWDL 301
L + D + WD +TG+ + C ++ L S + SS +L+ W
Sbjct: 103 LASSSVDDGIGCWDLHTGAEQLRYNSCSSPFHGLVSLGSRFLASSQLHHPSSASLFFWSF 282
Query: 302 NSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCT---NTIIFASN 358
+ +V + + + ++V A I +W++ G + + A
Sbjct: 283 SKPQVE--VKSFPAEAIKPLAANHQGTYLVGGALSGDIYLWEVETGRLLKKWHAHLRAVT 456
Query: 359 CNALCFSMDGQTIYSGHVDGNLRLWDIQS---------GKLLSEVA--AHSLAVTSISLS 407
C L FS D + SG DG +R+W + S K L E + H++ VT + +
Sbjct: 457 C--LVFSDDDSLLISGSEDGYVRVWSLFSLFDDLRSREEKNLYEYSFSEHTMRVTDVVIG 630
Query: 408 RNG-NVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNR---VASNWSRSCISPDNSHVAAGS 463
G N ++ S D+ R+ +V S R T R S + + P AGS
Sbjct: 631 NGGCNAIIVSASDD-------RTCKVWSLSRGTLLRSIVFPSVINAIVLDPAEHVFYAGS 789
Query: 464 ADGSVHIWSVHKNEI----------VSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
+DG + I +++ I + + H+ +V C +S L S ++GIV +W
Sbjct: 790 SDGKIFIAALNTERITTIDNYGMHIIGSFSNHSKAVTCLAYSKGRNLLISGSEDGIVRVW 969
>TC77248 similar to GP|13489182|gb|AAK27816.1 putative WD-repeat containing
protein {Oryza sativa}, partial (92%)
Length = 2005
Score = 65.9 bits (159), Expect = 4e-11
Identities = 67/286 (23%), Positives = 119/286 (41%), Gaps = 53/286 (18%)
Frame = +1
Query: 275 VLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAA 334
++ L I H + + N ++D SG+V TLTGH KV +V +++ +
Sbjct: 769 IIALDILHSKDLIATGNIDTNAVIFDRPSGQVLATLTGHSKKVTSVKFVG-QGESIITGS 945
Query: 335 YDRTIKVW---------------------------------------------DLMKGYC 349
D+T+++W +L G C
Sbjct: 946 ADKTVRLWQGSDDGHYNCKQILKDHSAEVEAVTVHATNNYFVTASLDGSWCFYELSSGTC 1125
Query: 350 TNTIIFAS-NCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSR 408
+ +S + F DG + +G D +++WD++S +++ H VT+IS S
Sbjct: 1126 LTQVSDSSEGYTSAAFHPDGLILGTGTTDSLVKIWDVKSQANVAKFDGHVGHVTAISFSE 1305
Query: 409 NGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSC-ISPDNSHVAAGSADGS 467
NG + T+ D V L+D+R L+ +FRD G ++ + S S++A G +D
Sbjct: 1306 NGYYLATAAHDGV-KLWDLRKLK---NFRDYGPYDSATPTNSVEFDHSGSYLAVGGSDVR 1473
Query: 468 VHIWSVHKNE--IVSTLKE--HTSSVLCCRWSGIGKPL--ASADKN 507
+ + K+E ++ TL + T C ++ K L AS D+N
Sbjct: 1474 ILQVANVKSEWNLIKTLPDLSGTGKSTCVKFGPDSKYLAVASMDRN 1611
Score = 37.4 bits (85), Expect = 0.014
Identities = 21/81 (25%), Positives = 40/81 (48%)
Frame = +1
Query: 221 TCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTI 280
TC +++ G S F + L TG D VK+WD + + + G +G V ++
Sbjct: 1120 TCLTQVSDSSEGYTSAAFHPDGLILGTGTTDSLVKIWDVKSQANVAKFDGHVGHVTAISF 1299
Query: 281 THDNRSVIAASSSNNLYVWDL 301
+ +N +A ++ + + +WDL
Sbjct: 1300 S-ENGYYLATAAHDGVKLWDL 1359
>TC78145 similar to GP|15294240|gb|AAK95297.1 AT3g18860/MCB22_3 {Arabidopsis
thaliana}, partial (93%)
Length = 2633
Score = 65.9 bits (159), Expect = 4e-11
Identities = 63/245 (25%), Positives = 110/245 (44%), Gaps = 6/245 (2%)
Frame = +3
Query: 245 LITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSG 304
+++GG D V VWD N+G +L G V + + D+ ++++S L W +G
Sbjct: 336 VVSGGMDTLVLVWDLNSGEKVQSLKGHQLQVTGIVV--DDGDLVSSSMDCTLRRW--RNG 503
Query: 305 RVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGY-CTNTII-FASNCNAL 362
+ T HK + AV K+ + +V+ + D T+K W KG C +T A L
Sbjct: 504 QCIETWEAHKGPIQAV--IKLPTGELVTGSSDTTLKTW---KGKTCLHTFQGHADTVRGL 668
Query: 363 CFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVH 422
D + + H DG+LRLW + SG+ L E+ H+ V S+ +G ++++ D
Sbjct: 669 AVMSDLGILSASH-DGSLRLWAV-SGEGLMEMVGHTAIVYSVDSHASG-LIVSGSEDRFA 839
Query: 423 NLFD----VRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEI 478
++ V+S+E DT +N + +DG IW+V+++
Sbjct: 840 KIWKDGVCVQSIEHPGCVWDTKFM------------ENGDIVTACSDGIARIWTVNQDYF 983
Query: 479 VSTLK 483
L+
Sbjct: 984 ADQLE 998
Score = 42.0 bits (97), Expect = 6e-04
Identities = 51/256 (19%), Positives = 101/256 (38%), Gaps = 41/256 (16%)
Frame = +3
Query: 300 DLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVW--DLMKGYCTNTII-FA 356
D ++R L GH+D V + + + + +++ D+TI++W D K ++
Sbjct: 99 DFKEYQLRCELHGHEDDVRGITICGGDNGGIATSSRDKTIRLWTRDNRKFVLDKVLVGHT 278
Query: 357 SNCNALCF-----SMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLS---- 407
S + + + + SG +D + +WD+ SG+ + + H L VT I +
Sbjct: 279 SFVGPIAWIPPNPDLPHGGVVSGGMDTLVLVWDLNSGEKVQSLKGHQLQVTGIVVDDGDL 458
Query: 408 ------------RNGNVVLT--SGRDNVHNLFDVRSLEVCSSFRDTGNRVASNW-SRSCI 452
RNG + T + + + + + + E+ + DT W ++C+
Sbjct: 459 VSSSMDCTLRRWRNGQCIETWEAHKGPIQAVIKLPTGELVTGSSDT---TLKTWKGKTCL 629
Query: 453 SPDNSH--------------VAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIG 498
H + + S DGS+ +W+V E + + HT+ V G
Sbjct: 630 HTFQGHADTVRGLAVMSDLGILSASHDGSLRLWAV-SGEGLMEMVGHTAIVYSVDSHASG 806
Query: 499 KPLASADKNGIVCIWK 514
+ S ++ IWK
Sbjct: 807 L-IVSGSEDRFAKIWK 851
>TC78016 similar to GP|21537191|gb|AAM61532.1 PRL1 protein {Arabidopsis
thaliana}, partial (81%)
Length = 1759
Score = 50.1 bits (118), Expect(2) = 7e-11
Identities = 24/88 (27%), Positives = 44/88 (49%)
Frame = +1
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNR 285
++ H G S+ + +++ TG D+++K+WD +G + L G I V L I+H +
Sbjct: 574 ISGHLGWVRSVAVDPSNTWFATGSADRTIKIWDLASGVLKLTLTGHIEQVRGLAISHKHT 753
Query: 286 SVIAASSSNNLYVWDLNSGRVRHTLTGH 313
+ +A + WDL +V + GH
Sbjct: 754 YMFSAGDDKQVKCWDLEQNKVIRSYHGH 837
Score = 46.2 bits (108), Expect = 3e-05
Identities = 24/77 (31%), Positives = 41/77 (53%)
Frame = +1
Query: 268 LHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSS 327
+ G +G V + + N S+ + +WDL SG ++ TLTGH ++V + +S
Sbjct: 574 ISGHLGWVRSVAVDPSNTWFATGSADRTIKIWDLASGVLKLTLTGHIEQVRGLAISH-KH 750
Query: 328 RHVVSAAYDRTIKVWDL 344
++ SA D+ +K WDL
Sbjct: 751 TYMFSAGDDKQVKCWDL 801
Score = 42.0 bits (97), Expect(2) = 1e-08
Identities = 27/107 (25%), Positives = 49/107 (45%), Gaps = 1/107 (0%)
Frame = +1
Query: 310 LTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTII-FASNCNALCFSMDG 368
++GH V +V V ++ +A DRTIK+WDL G T+ L S
Sbjct: 574 ISGHLGWVRSVAVDPSNTWFATGSA-DRTIKIWDLASGVLKLTLTGHIEQVRGLAISHKH 750
Query: 369 QTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLT 415
++S D ++ WD++ K++ H V +++ +++LT
Sbjct: 751 TYMFSAGDDKQVKCWDLEQNKVIRSYHGHLSGVYCLAIHPTIDILLT 891
Score = 41.2 bits (95), Expect = 0.001
Identities = 22/92 (23%), Positives = 40/92 (42%)
Frame = +3
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNR 285
L+ HE S+ +++TG D ++K+WD G S L SV + H
Sbjct: 954 LSGHENTVCSVFTRPTDPQVVTGSHDSTIKMWDLRYGKTMSTLTNHKKSVRAMA-QHPKE 1130
Query: 286 SVIAASSSNNLYVWDLNSGRVRHTLTGHKDKV 317
A++S++N+ + L G H + + +
Sbjct: 1131 QAFASASADNIKKFTLPKGEFCHNMLSQQKTI 1226
Score = 40.0 bits (92), Expect = 0.002
Identities = 28/102 (27%), Positives = 45/102 (43%), Gaps = 1/102 (0%)
Frame = +3
Query: 253 SVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTG 312
S VWD + L G +V + + V+ S + + +WDL G+ TLT
Sbjct: 909 SAGVWDIRSKMQIHALSGHENTVCSVFTRPTDPQVVTGSHDSTIKMWDLRYGKTMSTLTN 1088
Query: 313 HKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKG-YCTNTI 353
HK V A +++ ++A IK + L KG +C N +
Sbjct: 1089 HKKSVRA--MAQHPKEQAFASASADNIKKFTLPKGEFCHNML 1208
Score = 35.4 bits (80), Expect(2) = 1e-08
Identities = 26/99 (26%), Positives = 45/99 (45%)
Frame = +3
Query: 411 NVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHI 470
+V+L++G ++D+RS + N V S ++R P + V GS D ++ +
Sbjct: 897 DVILSAG------VWDIRSKMQIHALSGHENTVCSVFTR----PTDPQVVTGSHDSTIKM 1046
Query: 471 WSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGI 509
W + + +STL H SV + ASA + I
Sbjct: 1047WDLRYGKTMSTLTNHKKSVRAMAQHPKEQAFASASADNI 1163
Score = 34.7 bits (78), Expect(2) = 7e-11
Identities = 18/83 (21%), Positives = 37/83 (43%), Gaps = 2/83 (2%)
Frame = +3
Query: 341 VWDLMKGYCTNTIIFASN--CNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHS 398
VWD+ + + N C+ D Q + H D +++WD++ GK +S + H
Sbjct: 918 VWDIRSKMQIHALSGHENTVCSVFTRPTDPQVVTGSH-DSTIKMWDLRYGKTMSTLTNHK 1094
Query: 399 LAVTSISLSRNGNVVLTSGRDNV 421
+V +++ ++ DN+
Sbjct: 1095KSVRAMAQHPKEQAFASASADNI 1163
Score = 32.3 bits (72), Expect = 0.45
Identities = 16/44 (36%), Positives = 22/44 (49%), Gaps = 1/44 (2%)
Frame = +1
Query: 447 WSRS-CISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSV 489
W RS + P N+ A GSAD ++ IW + + TL H V
Sbjct: 592 WVRSVAVDPSNTWFATGSADRTIKIWDLASGVLKLTLTGHIEQV 723
Score = 30.4 bits (67), Expect = 1.7
Identities = 15/58 (25%), Positives = 22/58 (37%)
Frame = +1
Query: 223 KFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTI 280
K L H + + + + G D+ VK WD V + HG + V L I
Sbjct: 691 KLTLTGHIEQVRGLAISHKHTYMFSAGDDKQVKCWDLEQNKVIRSYHGHLSGVYCLAI 864
>TC78038 weakly similar to GP|21592898|gb|AAM64848.1 unknown {Arabidopsis
thaliana}, partial (60%)
Length = 1986
Score = 62.0 bits (149), Expect = 5e-10
Identities = 46/214 (21%), Positives = 90/214 (41%), Gaps = 21/214 (9%)
Frame = +3
Query: 245 LITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDL--- 301
L+ GG + +W+ TG + HG +V L + D+ +I+ + ++ VW L
Sbjct: 456 LVGGGLSGEIYLWEVETGRLLKKWHGHNTAVTCLVFSEDDSLLISGFKNGSVRVWSLLMI 635
Query: 302 --------NSGRVRHTLTGHKDKVCAVDVSKVSSRHVV-SAAYDRTIKVWDLMKGYCTNT 352
S ++ + H V V + ++ SA+ DRT KVW L G
Sbjct: 636 FDDVRRREASKIYEYSFSDHTLCVNDVVIGYGGCNAIIASASDDRTCKVWTLSNGMLQRN 815
Query: 353 IIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLS---------EVAAHSLAVTS 403
I+F S A+ +Y+G DG + + + + ++ + + HS AVT
Sbjct: 816 IVFPSKIKAIALDPAEHVLYAGSEDGKIFVAALNTTRITTNDQVSYITGSFSNHSKAVTC 995
Query: 404 ISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFR 437
++ S N +++ D + +++ + + F+
Sbjct: 996 LAYSAAENFLISGSDDGIVRVWNASTHNIIRVFK 1097
Score = 49.7 bits (117), Expect = 3e-06
Identities = 64/294 (21%), Positives = 116/294 (38%), Gaps = 37/294 (12%)
Frame = +3
Query: 257 WDTNTGS-------VCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHT 309
WD NTGS S HG + SV D + ++ A++SS +++ W + +V
Sbjct: 228 WDLNTGSEQIRYKSFSSPSHGLV-SVGDRFLA-SSQLRDASTSSASVFFWSWSKPQVE-- 395
Query: 310 LTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWD------LMKGYCTNTIIFASNCNALC 363
+ + S S ++V I +W+ L K + NT + L
Sbjct: 396 VKSFPSEPITPLASNHSGTYLVGGGLSGEIYLWEVETGRLLKKWHGHNTAV-----TCLV 560
Query: 364 FSMDGQTIYSGHVDGNLRLWDI----------QSGKLLS-EVAAHSLAVTSISLSRNG-N 411
FS D + SG +G++R+W + ++ K+ + H+L V + + G N
Sbjct: 561 FSEDDSLLISGFKNGSVRVWSLLMIFDDVRRREASKIYEYSFSDHTLCVNDVVIGYGGCN 740
Query: 412 VVLTSGRDNVHNLFDVRSLEVCSSFRDTGNR---VASNWSRSCISPDNSHVAAGSADGSV 468
++ S D+ R+ +V + R S + P + AGS DG +
Sbjct: 741 AIIASASDD-------RTCKVWTLSNGMLQRNIVFPSKIKAIALDPAEHVLYAGSEDGKI 899
Query: 469 HIWSVHKNEIVS---------TLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
+ +++ I + + H+ +V C +S L S +GIV +W
Sbjct: 900 FVAALNTTRITTNDQVSYITGSFSNHSKAVTCLAYSAAENFLISGSDDGIVRVW 1061
Score = 47.0 bits (110), Expect = 2e-05
Identities = 44/236 (18%), Positives = 95/236 (39%), Gaps = 27/236 (11%)
Frame = +3
Query: 275 VLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAA 334
+ L H ++ S +Y+W++ +GR+ GH V + S+ S ++S
Sbjct: 420 ITPLASNHSGTYLVGGGLSGEIYLWEVETGRLLKKWHGHNTAVTCLVFSEDDSL-LISGF 596
Query: 335 YDRTIKVWDLMKGY---------------------CTNTIIFA-SNCNALCFSMDGQTIY 372
+ +++VW L+ + C N ++ CNA+ I
Sbjct: 597 KNGSVRVWSLLMIFDDVRRREASKIYEYSFSDHTLCVNDVVIGYGGCNAI--------IA 752
Query: 373 SGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEV 432
S D ++W + +G L + S + +I+L +V+ D + + + +
Sbjct: 753 SASDDRTCKVWTLSNGMLQRNIVFPS-KIKAIALDPAEHVLYAGSEDGKIFVAALNTTRI 929
Query: 433 CSSFR---DTGNRVASNWSRSCI--SPDNSHVAAGSADGSVHIWSVHKNEIVSTLK 483
++ + TG+ + + +C+ S + + +GS DG V +W+ + I+ K
Sbjct: 930 TTNDQVSYITGSFSNHSKAVTCLAYSAAENFLISGSDDGIVRVWNASTHNIIRVFK 1097
Score = 37.0 bits (84), Expect = 0.018
Identities = 36/150 (24%), Positives = 59/150 (39%), Gaps = 9/150 (6%)
Frame = +3
Query: 373 SGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSG--RDNVHNLFDV--- 427
S G + WD+ +G ++ S + S L G+ L S RD + V
Sbjct: 198 STDASGGIGCWDLNTGS--EQIRYKSFSSPSHGLVSVGDRFLASSQLRDASTSSASVFFW 371
Query: 428 ----RSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLK 483
+EV S + +ASN S +++ G G +++W V ++
Sbjct: 372 SWSKPQVEVKSFPSEPITPLASNHS-------GTYLVGGGLSGEIYLWEVETGRLLKKWH 530
Query: 484 EHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
H ++V C +S L S KNG V +W
Sbjct: 531 GHNTAVTCLVFSEDDSLLISGFKNGSVRVW 620
>TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {Arabidopsis
thaliana}, partial (44%)
Length = 1098
Score = 61.2 bits (147), Expect = 9e-10
Identities = 51/225 (22%), Positives = 95/225 (41%), Gaps = 10/225 (4%)
Frame = +2
Query: 298 VWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTII-FA 356
VWD+ + + H +DV ++ S++ D I V + + + T T
Sbjct: 146 VWDVKAEECKQQFGFHSGPT--LDVDWRTNTSFASSSTDNMIYVCKIGENHPTQTFAGHQ 319
Query: 357 SNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGN----- 411
N + + G + S D ++W ++ K L + HS + +I S G
Sbjct: 320 GEVNCVKWDPTGTLLASCSDDITAKIWSVKQDKYLHDFREHSKEIYTIRWSPTGPGTNNP 499
Query: 412 ----VVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGS 467
++ ++ D+ L+D+ ++ S + V S SP+ ++A+GS D S
Sbjct: 500 NKKLLLASASFDSTVKLWDIELGKLIHSLNGHRHPVYS----VAFSPNGEYIASGSLDKS 667
Query: 468 VHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCI 512
+HIWS+ + +I+ T C W+ G +A+ N IVC+
Sbjct: 668 LHIWSLKEGKIIRTYNGSGGIFEVC-WNKEGDKIAACFANNIVCV 799
Score = 55.8 bits (133), Expect = 4e-08
Identities = 43/204 (21%), Positives = 83/204 (40%), Gaps = 10/204 (4%)
Frame = +2
Query: 256 VWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKD 315
VWD G LD+ N S ++S+ N +YV + T GH+
Sbjct: 146 VWDVKAEECKQQFGFHSGPTLDVD-WRTNTSFASSSTDNMIYVCKIGENHPTQTFAGHQG 322
Query: 316 KVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKG-YCTNTIIFASNCNALCFSMDGQ----- 369
+V V + + S + D T K+W + + Y + + + +S G
Sbjct: 323 EVNCVKWDPTGTL-LASCSDDITAKIWSVKQDKYLHDFREHSKEIYTIRWSPTGPGTNNP 499
Query: 370 ----TIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLF 425
+ S D ++LWDI+ GKL+ + H V S++ S NG + + D +++
Sbjct: 500 NKKLLLASASFDSTVKLWDIELGKLIHSLNGHRHPVYSVAFSPNGEYIASGSLDKSLHIW 679
Query: 426 DVRSLEVCSSFRDTGNRVASNWSR 449
++ ++ ++ +G W++
Sbjct: 680 SLKEGKIIRTYNGSGGIFEVCWNK 751
Score = 35.0 bits (79), Expect = 0.069
Identities = 16/58 (27%), Positives = 25/58 (42%)
Frame = +2
Query: 456 NSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
N+ A+ S D +++ + +N T H V C +W G LAS + IW
Sbjct: 227 NTSFASSSTDNMIYVCKIGENHPTQTFAGHQGEVNCVKWDPTGTLLASCSDDITAKIW 400
Score = 33.9 bits (76), Expect = 0.15
Identities = 21/92 (22%), Positives = 38/92 (40%), Gaps = 2/92 (2%)
Frame = +2
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNR 285
LN H S+ F N + +G D+S+ +W G + +G G + ++ +
Sbjct: 584 LNGHRHPVYSVAFSPNGEYIASGSLDKSLHIWSLKEGKIIRTYNGS-GGIFEVCWNKEGD 760
Query: 286 SVIAASSSNNLYVWDLNS--GRVRHTLTGHKD 315
+ A ++N + V D + R L G D
Sbjct: 761 KIAACFANNIVCVLDFRM*LWKCRVVLAGTSD 856
>TC82910 similar to GP|19347844|gb|AAL86002.1 unknown protein {Arabidopsis
thaliana}, partial (19%)
Length = 821
Score = 60.5 bits (145), Expect = 2e-09
Identities = 42/174 (24%), Positives = 78/174 (44%), Gaps = 1/174 (0%)
Frame = +1
Query: 264 VCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVS 323
+C +H + L I + + S ++++ ++ +GRV TL H + V +
Sbjct: 19 LCEVIHTIPRGITCLAINSTSTIALTGSVDGSVHIVNITTGRVVSTLPSHSSSIECVGFA 198
Query: 324 KVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALCFSMDGQT-IYSGHVDGNLRL 382
S + TI WD+ + I C + G + + +G DG +RL
Sbjct: 199 PSGSWAAIGGMDKMTI--WDVEHSLARS--ICEHEYGVTCSTWLGTSYVATGSNDGAVRL 366
Query: 383 WDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSF 436
WD +SG+ + HS ++ S+SLS N ++++ D+ +FDV+ + SF
Sbjct: 367 WDSRSGECVRTFRGHSESIQSLSLSANQEYLVSASLDHTARVFDVKGVC*AKSF 528
Score = 54.7 bits (130), Expect = 8e-08
Identities = 35/132 (26%), Positives = 63/132 (47%)
Frame = +1
Query: 373 SGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEV 432
+G VDG++ + +I +G+++S + +HS ++ + + +G+ G D + ++DV
Sbjct: 94 TGSVDGSVHIVNITTGRVVSTLPSHSSSIECVGFAPSGSWAAIGGMDKM-TIWDVEHSLA 270
Query: 433 CSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCC 492
S S W + S+VA GS DG+V +W E V T + H+ S+
Sbjct: 271 RSICEHEYGVTCSTWLGT------SYVATGSNDGAVRLWDSRSGECVRTFRGHSESIQSL 432
Query: 493 RWSGIGKPLASA 504
S + L SA
Sbjct: 433 SLSANQEYLVSA 468
Score = 39.3 bits (90), Expect = 0.004
Identities = 21/57 (36%), Positives = 33/57 (57%), Gaps = 8/57 (14%)
Frame = +1
Query: 450 SCISPDNSHVAA--GSADGSVHIWSVHKNEIVSTLKEHTSSVLCC------RWSGIG 498
+C++ +++ A GS DGSVHI ++ +VSTL H+SS+ C W+ IG
Sbjct: 55 TCLAINSTSTIALTGSVDGSVHIVNITTGRVVSTLPSHSSSIECVGFAPSGSWAAIG 225
>BG457241 homologue to PIR|F84600|F8 coatomer alpha subunit [imported] -
Arabidopsis thaliana, partial (15%)
Length = 654
Score = 59.7 bits (143), Expect = 3e-09
Identities = 33/121 (27%), Positives = 59/121 (48%), Gaps = 1/121 (0%)
Frame = +1
Query: 225 RLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDN 284
+ + H+G + F + ++GG D +KVW+ L G + + + H++
Sbjct: 208 KFDEHDGPVRGVHFHNSQPLFVSGGDDYKIKVWNYKLHRCLFTLLGHLDYIRTVQFHHES 387
Query: 285 RSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKV-CAVDVSKVSSRHVVSAAYDRTIKVWD 343
+++AS + +W+ S LTGH V CA+ K VVSA+ D+T++VWD
Sbjct: 388 PWIVSASDDQTIRIWNWQSRTCISVLTGHNHYVMCALFHPK--DDLVVSASLDQTVRVWD 561
Query: 344 L 344
+
Sbjct: 562 I 564
Score = 52.4 bits (124), Expect = 4e-07
Identities = 36/148 (24%), Positives = 63/148 (42%), Gaps = 2/148 (1%)
Frame = +1
Query: 254 VKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGH 313
+++WD G++ G V + + ++ + VW+ R TL GH
Sbjct: 169 IQLWDYRMGTLIDKFDEHDGPVRGVHFHNSQPLFVSGGDDYKIKVWNYKLHRCLFTLLGH 348
Query: 314 KDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALC--FSMDGQTI 371
D + V S +VSA+ D+TI++W+ C +++ N +C F +
Sbjct: 349 LDYIRTVQFHH-ESPWIVSASDDQTIRIWNWQSRTCI-SVLTGHNHYVMCALFHPKDDLV 522
Query: 372 YSGHVDGNLRLWDIQSGKLLSEVAAHSL 399
S +D +R+WDI S K S A +
Sbjct: 523 VSASLDQTVRVWDIGSLKRKSASXADDI 606
Score = 37.4 bits (85), Expect = 0.014
Identities = 20/82 (24%), Positives = 38/82 (45%)
Frame = +1
Query: 222 CKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTIT 281
C F L H ++ F S +++ DQ++++W+ + + S L G V+
Sbjct: 325 CLFTLLGHLDYIRTVQFHHESPWIVSASDDQTIRIWNWQSRTCISVLTGHNHYVMCALFH 504
Query: 282 HDNRSVIAASSSNNLYVWDLNS 303
+ V++AS + VWD+ S
Sbjct: 505 PKDDLVVSASLDQTVRVWDIGS 570
Score = 33.1 bits (74), Expect = 0.26
Identities = 28/137 (20%), Positives = 59/137 (42%), Gaps = 1/137 (0%)
Frame = +1
Query: 378 GNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFR 437
G ++LWD + G L+ + H V + + + ++ G D +++ + L C F
Sbjct: 163 GVIQLWDYRMGTLIDKFDEHDGPVRGVHFHNSQPLFVSGGDDYKIKVWNYK-LHRCL-FT 336
Query: 438 DTGNRVASNWSRSC-ISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSG 496
G+ ++ R+ ++ + + S D ++ IW+ +S L H V+C +
Sbjct: 337 LLGHL---DYIRTVQFHHESPWIVSASDDQTIRIWNWQSRTCISVLTGHNHYVMCALFHP 507
Query: 497 IGKPLASADKNGIVCIW 513
+ SA + V +W
Sbjct: 508 KDDLVVSASLDQTVRVW 558
>TC78442 homologue to GP|5733806|gb|AAD49742.1| G protein beta subunit
{Pisum sativum}, complete
Length = 1311
Score = 50.8 bits (120), Expect(2) = 3e-09
Identities = 42/177 (23%), Positives = 72/177 (39%), Gaps = 14/177 (7%)
Frame = +2
Query: 222 CKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTIT 281
C L H G S+ + +R+++ QD + VW+ T + V+ +
Sbjct: 299 CCRTLQGHTGKVYSLDWTSEKNRIVSASQDGRLIVWNALTSQKTHAIKLPCAWVMTCAFS 478
Query: 282 HDNRSVIAASSSNNLYVWDLNSG-------RVRHTLTGHKDKVCAVDVSKVSSRHVVSAA 334
+SV + +++LNS V L+GHK V + H+++ +
Sbjct: 479 PTGQSVACGGLDSVCSIFNLNSPTDRDGNLNVSRMLSGHKGYVSSCQYVPGEDTHLITGS 658
Query: 335 YDRTIKVWDLMKGYCTNTI--IFAS--NCNALCFSMDG---QTIYSGHVDGNLRLWD 384
D+T +WD+ G T+ F S + L S++G + SG D RLWD
Sbjct: 659 GDQTCVLWDITTGLRTSVFGGEFQSGHTADVLSISINGSNSKMFVSGSCDATARLWD 829
Score = 28.5 bits (62), Expect(2) = 3e-09
Identities = 26/119 (21%), Positives = 45/119 (36%), Gaps = 7/119 (5%)
Frame = +3
Query: 403 SISLSRNGNVVLTSGRDNVHNLFDVRS---LEVCSSFRDTGNRVASNWSRSCISPDNSHV 459
S+ +GN T D LFD+R+ L+V + + +G+ ++ + S +
Sbjct: 888 SVKFFPDGNRFGTGSEDGTCRLFDIRTGHQLQVYNQ-QHSGDNEMAHVTSIAFSISGRLL 1064
Query: 460 AAGSADGSVHIWSVHKNEIVSTL----KEHTSSVLCCRWSGIGKPLASADKNGIVCIWK 514
AG +G ++W ++V L H + C G VC WK
Sbjct: 1065IAGYTNGDCYVWDTLLAKVVLNLGSLQNSHEGRITCL---------------GFVC*WK 1196
>BG648991 similar to PIR|C86239|C86 protein T10O24.21 [imported] -
Arabidopsis thaliana, partial (35%)
Length = 702
Score = 59.3 bits (142), Expect = 3e-09
Identities = 40/136 (29%), Positives = 61/136 (44%), Gaps = 3/136 (2%)
Frame = +3
Query: 218 IPSTCKFRLNAHEGGCASILFEFNSSRLI-TGGQDQSVKVWDT-NTGSVCSNLHGCIGSV 275
IP + H G ++I F NS LI + G D VK+WD NTG G +V
Sbjct: 291 IPKRLVHTWSGHTKGVSAIRFFPNSGHLILSAGMDTKVKIWDVFNTGKCMRTYMGHSKAV 470
Query: 276 LDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTL-TGHKDKVCAVDVSKVSSRHVVSAA 334
D+ T+D ++A N+ WD +G+V T TG V ++ + +++
Sbjct: 471 RDICFTNDGTKFLSAGYDKNIKYWDTETGQVISTFSTGKIPYVVKLNPDEDKQNVLLAGM 650
Query: 335 YDRTIKVWDLMKGYCT 350
D+ I WD+ G T
Sbjct: 651 SDKKIVQWDMNTGQIT 698
Score = 56.6 bits (135), Expect = 2e-08
Identities = 32/106 (30%), Positives = 55/106 (51%), Gaps = 2/106 (1%)
Frame = +3
Query: 289 AASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMK-G 347
A +S+++ Y+ R+ HT +GH V A+ S ++SA D +K+WD+ G
Sbjct: 264 AKASNDHCYI----PKRLVHTWSGHTKGVSAIRFFPNSGHLILSAGMDTKVKIWDVFNTG 431
Query: 348 YCTNTIIFASNC-NALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLS 392
C T + S +CF+ DG S D N++ WD ++G+++S
Sbjct: 432 KCMRTYMGHSKAVRDICFTNDGTKFLSAGYDKNIKYWDTETGQVIS 569
Score = 40.4 bits (93), Expect = 0.002
Identities = 25/108 (23%), Positives = 49/108 (45%), Gaps = 4/108 (3%)
Frame = +3
Query: 287 VIAASSSNNLYVWDL-NSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLM 345
+++A + +WD+ N+G+ T GH V + + ++ + SA YD+ IK WD
Sbjct: 375 ILSAGMDTKVKIWDVFNTGKCMRTYMGHSKAVRDICFTNDGTKFL-SAGYDKNIKYWDTE 551
Query: 346 KGYCTNTIIFASNCNALCFSMD---GQTIYSGHVDGNLRLWDIQSGKL 390
G +T + + D + +G D + WD+ +G++
Sbjct: 552 TGQVISTFSTGKIPYVVKLNPDEDKQNVLLAGMSDKKIVQWDMNTGQI 695
>TC83414 weakly similar to GP|17381206|gb|AAL36415.1 unknown protein
{Arabidopsis thaliana}, partial (28%)
Length = 1088
Score = 58.5 bits (140), Expect = 6e-09
Identities = 62/307 (20%), Positives = 131/307 (42%), Gaps = 31/307 (10%)
Frame = +2
Query: 238 FEFNSSRLITGGQDQSVKVW--------------DTNTGSVCSNLHGCIGSVLDLTITHD 283
F + L +GG+D V+VW D + ++ ++ G V L + D
Sbjct: 5 FSLDGKYLASGGEDGIVRVWQVVENERSNELNILDNDPSNIYFKMNQFTGCVAPLDVDKD 184
Query: 284 ---NRSVIAASSSNNLYVWDLNSGRV----RHTLTGHKDKVCAVDVSKVSSRHVVSAAYD 336
+ SS++ + + R+ H GH D + +D++ S ++S++ D
Sbjct: 185 KLVKTEKLRKSSASTCVIIPPKTFRILVKPLHEFQGHSDDI--LDLAWSKSGFLLSSSVD 358
Query: 337 RTIKVWDLMKGYCTNTIIFASNCNALCFS---MDGQTIYSGHVDGNLRLWDIQSGKLLSE 393
+T+++W + C +F+ N C + ++ SG +DG +R+W++ +++
Sbjct: 359 KTVRLWQVGISKCLR--VFSHNNYVTCVNFNPVNDNFFISGSIDGKVRIWEVVRCRVVDY 532
Query: 394 VAAHSLAVTSISLSRNGN---VVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVAS--NWS 448
+ + VT++ NG V +G +N+ D +++ + F G + S +
Sbjct: 533 IDIREI-VTAVCFCPNGQGTIVGTMTGNCRFYNILD-NHMKMDAEFSLQGKKKTSGKRIT 706
Query: 449 RSCISP-DNSHVAAGSADGSVHIWSVHKNEIVSTLKE-HTSSVLCCRWSGIGKPLASADK 506
SP D + + SAD H+ + +++ K ++ + ++ GK + S +
Sbjct: 707 GFQFSPSDPTKLLVASADS--HVCILSGVDVIYKFKGLRSAGQMHASFTSDGKHIVSLSE 880
Query: 507 NGIVCIW 513
VCIW
Sbjct: 881 GSNVCIW 901
Score = 48.9 bits (115), Expect = 5e-06
Identities = 63/301 (20%), Positives = 116/301 (37%), Gaps = 22/301 (7%)
Frame = +2
Query: 194 LARQQVDGVVR-----RCEEGAEFFLDSNIPSTCKFRLNAHEGGCASILFE----FNSSR 244
LA DG+VR E E + N PS F++N G A + + + +
Sbjct: 26 LASGGEDGIVRVWQVVENERSNELNILDNDPSNIYFKMNQFTGCVAPLDVDKDKLVKTEK 205
Query: 245 LITGGQDQSVKVWDTNTGSVCSNLH---GCIGSVLDLTITHDNRSVIAASSSNNLYVWDL 301
L V + + LH G +LDL + ++++S + +W +
Sbjct: 206 LRKSSASTCVIIPPKTFRILVKPLHEFQGHSDDILDLAWSKSG-FLLSSSVDKTVRLWQV 382
Query: 302 NSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNA 361
+ + H + V V+ + V+ +S + D +++W++++ + I A
Sbjct: 383 GISKCLRVFS-HNNYVTCVNFNPVNDNFFISGSIDGKVRIWEVVRCRVVDYIDIREIVTA 559
Query: 362 LCFSMDGQTIYSGHVDGNLRLWDIQSG--KLLSEVAAHSLAVTS--------ISLSRNGN 411
+CF +GQ G + GN R ++I K+ +E + TS S S
Sbjct: 560 VCFCPNGQGTIVGTMTGNCRFYNILDNHMKMDAEFSLQGKKKTSGKRITGFQFSPSDPTK 739
Query: 412 VVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIW 471
+++ S +V L V + R G AS + D H+ + S +V IW
Sbjct: 740 LLVASADSHVCILSGVDVIYKFKGLRSAGQMHAS------FTSDGKHIVSLSEGSNVCIW 901
Query: 472 S 472
+
Sbjct: 902 N 904
>TC82717 weakly similar to GP|21539583|gb|AAM53344.1 putative coatomer
complex subunit {Arabidopsis thaliana}, partial (29%)
Length = 1118
Score = 57.8 bits (138), Expect = 1e-08
Identities = 49/197 (24%), Positives = 86/197 (42%), Gaps = 4/197 (2%)
Frame = +1
Query: 251 DQSVKVWDTNTGSVCSN-LHGCIGSVLDLTITHDNRSVIAASS-SNNLYVWDLNSGRVRH 308
DQ +K+W+ G C G V+ + + S A++S L +W ++S
Sbjct: 448 DQVLKLWNWKKGWSCDETFEGNSHYVMQVAFNPKDPSTFASASLDGTLKIWTIDSSAPNF 627
Query: 309 TLTGHKDKVCAVDVSKVSSR-HVVSAAYDRTIKVWDLMKGYCTNTII-FASNCNALCFSM 366
T GH + VD + + + +++S + D T KVWD C T+ +N A+C
Sbjct: 628 TFEGHLKGMNCVDYFESNDKQYLLSGSDDYTAKVWDYDSKNCVQTLEGHKNNVTAICAHP 807
Query: 367 DGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFD 426
+ I + D +++WD + +L + + V SI + G+ L G D +
Sbjct: 808 EIPIIITASEDSTVKIWDAVTYRLQNTLDFGLERVWSIGY-KKGSSQLAFGCDKGFIIVT 984
Query: 427 VRSLEVCSSFRDTGNRV 443
V S C+ + D G R+
Sbjct: 985 VNSANYCTLY-DFGGRL 1032
Score = 48.1 bits (113), Expect = 8e-06
Identities = 28/115 (24%), Positives = 56/115 (48%), Gaps = 2/115 (1%)
Frame = +1
Query: 243 SRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSV--LDLTITHDNRSVIAASSSNNLYVWD 300
S + D ++K+W ++ + G + + +D ++D + +++ S VWD
Sbjct: 556 STFASASLDGTLKIWTIDSSAPNFTFEGHLKGMNCVDYFESNDKQYLLSGSDDYTAKVWD 735
Query: 301 LNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIF 355
+S TL GHK+ V A+ + +++A+ D T+K+WD + NT+ F
Sbjct: 736 YDSKNCVQTLEGHKNNVTAI-CAHPEIPIIITASEDSTVKIWDAVTYRLQNTLDF 897
Score = 44.7 bits (104), Expect = 9e-05
Identities = 34/141 (24%), Positives = 56/141 (39%), Gaps = 4/141 (2%)
Frame = +1
Query: 290 ASSSNNLYVWDLNSG-RVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGY 348
AS L +W+ G T G+ V V + SA+ D T+K+W +
Sbjct: 439 ASDDQVLKLWNWKKGWSCDETFEGNSHYVMQVAFNPKDPSTFASASLDGTLKIWTIDSSA 618
Query: 349 CTNTI---IFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSIS 405
T + NC S D Q + SG D ++WD S + + H VT+I
Sbjct: 619 PNFTFEGHLKGMNCVDYFESNDKQYLLSGSDDYTAKVWDYDSKNCVQTLEGHKNNVTAIC 798
Query: 406 LSRNGNVVLTSGRDNVHNLFD 426
+++T+ D+ ++D
Sbjct: 799 AHPEIPIIITASEDSTVKIWD 861
>TC91699 similar to GP|21592393|gb|AAM64344.1 unknown {Arabidopsis
thaliana}, partial (71%)
Length = 1035
Score = 57.4 bits (137), Expect = 1e-08
Identities = 45/209 (21%), Positives = 84/209 (39%), Gaps = 4/209 (1%)
Frame = +3
Query: 291 SSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCT 350
S+ +L + R GHK +V ++ +S ++ + S + D ++++WDL C
Sbjct: 36 STGESLRYLSMYDNRCLRYFKGHKQRVVSLCMSPINDSFM-SGSVDHSVRLWDLRVNACQ 212
Query: 351 NTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKL----LSEVAAHSLAVTSISL 406
I+ + + G G ++L+D +S V + V I
Sbjct: 213 G-ILHVRGRPTVAYDQQGLVFAVAMEGGAIKLFDSRSYDKGPFDTFLVGGDTAEVCDIKF 389
Query: 407 SRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADG 466
S +G +L S +N + D + F + S + +PD +V A S G
Sbjct: 390 SNDGKSMLLSTTNNNIYVLDAYGGDKRCGFSLEPSHGTS--IEATFTPDGKYVVASSGGG 563
Query: 467 SVHIWSVHKNEIVSTLKEHTSSVLCCRWS 495
++H WS+ +N V+ H C +W+
Sbjct: 564 TMHAWSIDRNHEVACWSSHIGVPSCLKWA 650
Score = 32.7 bits (73), Expect = 0.34
Identities = 19/94 (20%), Positives = 47/94 (49%), Gaps = 4/94 (4%)
Frame = +3
Query: 270 GCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVS-KVSSR 328
G V D+ ++D +S++ ++++NN+YV D G R + +++ + +
Sbjct: 357 GDTAEVCDIKFSNDGKSMLLSTTNNNIYVLDAYGGDKRCGFSLEPSHGTSIEATFTPDGK 536
Query: 329 HVVSAAYDRTIKVWDLMKGY---CTNTIIFASNC 359
+VV+++ T+ W + + + C ++ I +C
Sbjct: 537 YVVASSGGGTMHAWSIDRNHEVACWSSHIGVPSC 638
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.129 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,979,509
Number of Sequences: 36976
Number of extensions: 229421
Number of successful extensions: 1765
Number of sequences better than 10.0: 178
Number of HSP's better than 10.0 without gapping: 1380
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1623
length of query: 514
length of database: 9,014,727
effective HSP length: 100
effective length of query: 414
effective length of database: 5,317,127
effective search space: 2201290578
effective search space used: 2201290578
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0173b.8


