

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
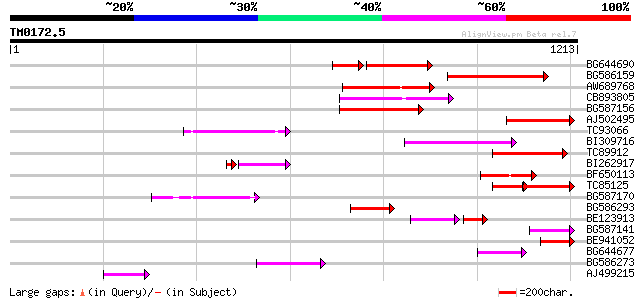
Query= TM0172.5
(1213 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 191 2e-70
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 191 2e-48
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 177 3e-44
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 174 2e-43
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 155 9e-38
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 149 6e-36
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 149 6e-36
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 147 2e-35
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 132 1e-30
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 112 4e-26
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 100 4e-21
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 96 1e-19
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 93 5e-19
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 82 1e-15
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 60 2e-15
BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440... 74 3e-13
BE941052 weakly similar to PIR|B85188|B85 retrotransposon like p... 72 1e-12
BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T... 65 2e-10
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 64 3e-10
AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 64 5e-10
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 191 bits (485), Expect(2) = 2e-70
Identities = 89/141 (63%), Positives = 114/141 (80%)
Frame = -2
Query: 763 VTRNKSRLVAQGYSQIEGIDFYETFAPVARLESIRLLLGVACLLKFRLYQMDVKSAFLNG 822
+TRNKS+LV QGY+Q EGID+ E F+PVAR+E+IR+L+ A + F+LYQMDVKSAF+NG
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 823 YLNEEVYVEQPKGFVDPSFPDHVYRLKKALYGLKQAPRAWYERLTEFLINNGYDKGGIDK 882
L EEV+V+QP GF D P+HV+RL K LYGLKQAPRAWYERL++FL+ NG+ +G ID
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 883 TLFVKKNGGELMVAQIYVDDI 903
TLF+ K EL++ Q+YVDDI
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 95.1 bits (235), Expect(2) = 2e-70
Identities = 43/65 (66%), Positives = 53/65 (81%)
Frame = -3
Query: 692 RDLVSNACFISKVEPKNVKEALTDEFWIQSMQEELGQFKRNEVWELVPRPDDANVVGTKW 751
R+LVS + FIS +EPKNVKEAL D WI SMQEEL QF+R++VW LVPRP+ V+GT+W
Sbjct: 627 RNLVSFSAFISSIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRW 448
Query: 752 IFRNK 756
+FRNK
Sbjct: 447 VFRNK 433
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 191 bits (484), Expect = 2e-48
Identities = 90/217 (41%), Positives = 140/217 (64%), Gaps = 1/217 (0%)
Frame = +1
Query: 938 LQVKQMEDTLFITQSKYAKGIVKKFGLENAGHKRTPAATHIKLTKDEKGTDVDPSLYRSM 997
++V Q E+ ++I Q KY ++++FG+E + R P A KL KDE G VD + Y+ +
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 998 IGSLLYLTASRPDITFAVGVCARYQAEPKTSHLIQVKRIIKYISGTSDYGILYSHNTNSG 1057
+G L+YL A+RPD+ + + + +R+ P H+ VKR+++Y++GT + GI+Y N +
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 1058 LTGYCDADWAGSADDRKSTSGGCFFLENNLISWFSKKQNCVSLSTAEAEYIAAGSSCTQL 1117
L Y D+D+AG DDRKSTSG F L + +SW SKKQ V+LST +AE+IAA Q
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 1118 MWMKQMLKEYN-VQQDVMTLFCDNLSAINISKNPIQH 1153
+WM+++L++ Q +T++CDN S I +SKNP+ H
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 177 bits (448), Expect = 3e-44
Identities = 88/197 (44%), Positives = 126/197 (63%)
Frame = +1
Query: 713 LTDEFWIQSMQEELGQFKRNEVWELVPRPDDANVVGTKWIFRNKSDESGNVTRNKSRLVA 772
L+D W+Q+M+ E N+ W+LVP P +G KW++R K + G+V + K+RLVA
Sbjct: 82 LSDPRWLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVA 261
Query: 773 QGYSQIEGIDFYETFAPVARLESIRLLLGVACLLKFRLYQMDVKSAFLNGYLNEEVYVEQ 832
+G+SQ G D+ ETF+PV + +IRL+L +A K+ + Q+D+ +AFLNG+L EEVY+ Q
Sbjct: 262 KGFSQTLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQ 441
Query: 833 PKGFVDPSFPDHVYRLKKALYGLKQAPRAWYERLTEFLINNGYDKGGIDKTLFVKKNGGE 892
P+GF + + V +L K+LYGLKQAPRAWYE LT I G+ K D +L + G
Sbjct: 442 PQGF-EAANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGA 618
Query: 893 LMVAQIYVDDIVFGGMS 909
+ IYVDDI+ G S
Sbjct: 619 CIYLXIYVDDILITGSS 669
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 174 bits (441), Expect = 2e-43
Identities = 94/252 (37%), Positives = 148/252 (58%), Gaps = 7/252 (2%)
Frame = +3
Query: 705 EPKNVKEALTDEFWIQSMQEELGQFKRNEVWELVPRPDDANVVGTKWIFRNKSDESGNVT 764
+P +EA+ E W SM E+ +RN WEL A +G KWIF+ K +E+G +
Sbjct: 39 DPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNENGEIE 218
Query: 765 RNKSRLVAQGYSQIEGIDFYETFAPVARLESIRLLLGVACLLK---FRLYQMDVKSAFLN 821
+ K+RLVA+GYSQ G+D+ E FAPVAR ++IR+++ +A +K + M + +
Sbjct: 219 KYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M*KAHSCME 398
Query: 822 GYLNEEVYVEQP---KGFVDPSFPDHVYRLKKALYGLKQAPRAWYERLTEFLINNGYDKG 878
+ + + + + + R+K+ALYGLKQAPRAWY R+ + G++K
Sbjct: 399 N*MRKFLLINHRVM*RRVIS*-------RVKRALYGLKQAPRAWYSRIEAYFTKEGFEKC 557
Query: 879 GIDKTLFVK-KNGGELMVAQIYVDDIVFGGMSNQMVEQFVEQMKSEFEMSLVGELTYFLG 937
+ TLFVK GG++++ +YVDD++F G M E+F + MK EF MS +G++ YFLG
Sbjct: 558 PYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMHYFLG 737
Query: 938 LQVKQMEDTLFI 949
++V Q E ++I
Sbjct: 738 VEVTQNEKGIYI 773
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 155 bits (392), Expect = 9e-38
Identities = 79/180 (43%), Positives = 114/180 (62%)
Frame = -1
Query: 706 PKNVKEALTDEFWIQSMQEELGQFKRNEVWELVPRPDDANVVGTKWIFRNKSDESGNVTR 765
P++ +EA+ D+ W +S+ E G +N+ W P V ++WIF K G++ R
Sbjct: 558 PRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYKADGSIER 379
Query: 766 NKSRLVAQGYSQIEGIDFYETFAPVARLESIRLLLGVACLLKFRLYQMDVKSAFLNGYLN 825
K+RLVA+G++ G D+ ETFAPVA+L +IR++L +A L + L+QMDVK+AFL G L
Sbjct: 378 KKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQGELE 199
Query: 826 EEVYVEQPKGFVDPSFPDHVYRLKKALYGLKQAPRAWYERLTEFLINNGYDKGGIDKTLF 885
+EVY+ P G +V RLKKA+YGLKQ+PRAWY +L+ L G+ K +D TLF
Sbjct: 198 DEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELDHTLF 19
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 149 bits (376), Expect = 6e-36
Identities = 74/146 (50%), Positives = 104/146 (70%), Gaps = 1/146 (0%)
Frame = +2
Query: 1064 ADWAGSADDRKSTSGGCFFLENNLISWFSKKQNCVSLSTAEAEYIAAGSSCTQLMWMKQM 1123
+DWAG + RKSTSG F L ISW SKKQ V+ STAEAEYIA+ S TQ +W++++
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 1124 LKEYNVQQDVMT-LFCDNLSAINISKNPIQHSRTKHIDIRHHFIRELVEDGTVTLEHVST 1182
L+ + +Q+ T ++CDN SAI +SKNP+ H R+KHIDI+ H IREL+ + V +E+ T
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 1183 EKQLADIFTKALDATQFEKLRQLLGI 1208
E+++ADIFTK L F KL+++LG+
Sbjct: 362 EEKIADIFTKPLKIESFYKLKKMLGM 439
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 149 bits (376), Expect = 6e-36
Identities = 85/230 (36%), Positives = 123/230 (52%), Gaps = 2/230 (0%)
Frame = +1
Query: 373 GKQVKMSHQKLQHLTTSKVLELLHMDLMGPMQVESLGGKRYVFVCVDDFSRFTWVEFLKE 432
G + K+S H T +L+ +H DL GP +V S GG+RY+ +DDF R WV FL+
Sbjct: 43 GNRKKVSFSTATHRTKG-ILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRY 219
Query: 433 KSDTFEVFKELCLQVQREKGSGIVKIRSDHGKEFENGQFSEFCSSEGIKHEFSAPITPQQ 492
K++TF FK+ + V+ + G + K+ +D+ EF + F+EFC++ GI + P PQQ
Sbjct: 220 KNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQ 399
Query: 493 NGVVERKNRTLQESARAMLHAKNL--PYHFWAEAINTACYIHNRVTIRQGDTVTQYELWK 550
NGV ER RTL E AR ML L W EA +TAC++ NR D ++W
Sbjct: 400 NGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWS 579
Query: 551 GKKPTVKYFHVFGSKCYILSDREHRRKLDPKSEVGIFLGYSGNSRAYRVY 600
G +FG Y L + KL P++ IFL Y+ S+ YR++
Sbjct: 580 GNLVDYSNLRIFGCPAYAL---VNDGKLAPRAGECIFLSYASESKGYRLW 720
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 147 bits (371), Expect = 2e-35
Identities = 81/239 (33%), Positives = 127/239 (52%)
Frame = +2
Query: 845 VYRLKKALYGLKQAPRAWYERLTEFLINNGYDKGGIDKTLFVKKNGGELMVAQIYVDDIV 904
V L+K++YGLKQA R WY +L+E LI+ GY + D +LF K +YVDDIV
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDIV 199
Query: 905 FGGMSNQMVEQFVEQMKSEFEMSLVGELTYFLGLQVKQMEDTLFITQSKYAKGIVKKFGL 964
G ++ + F++ +G L YFLGL+V + + + + Q KY +++ G
Sbjct: 200 LAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSGN 379
Query: 965 ENAGHKRTPAATHIKLTKDEKGTDVDPSLYRSMIGSLLYLTASRPDITFAVGVCARYQAE 1024
TP +KL + D + YR +IG L+YLT +RPDI+FAV +++ ++
Sbjct: 380 LAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVSK 559
Query: 1025 PKTSHLIQVKRIIKYISGTSDYGILYSHNTNSGLTGYCDADWAGSADDRKSTSGGCFFL 1083
P+ H R+++Y+ G+ YS +N L+ + D+DWA RKS +G FL
Sbjct: 560 PQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFL 736
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 132 bits (331), Expect = 1e-30
Identities = 64/162 (39%), Positives = 102/162 (62%), Gaps = 2/162 (1%)
Frame = +1
Query: 1033 VKRIIKYISGTSDYGILYSH--NTNSGLTGYCDADWAGSADDRKSTSGGCFFLENNLISW 1090
+K ++KY++ + + Y+ L GY DAD+AG+ D RKS SG F L ISW
Sbjct: 7 LKWVLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTISW 186
Query: 1091 FSKKQNCVSLSTAEAEYIAAGSSCTQLMWMKQMLKEYNVQQDVMTLFCDNLSAINISKNP 1150
+ +Q+ V+LST +AEYIA +W+K M+ E + Q+ + + CD+ SAI+++ +
Sbjct: 187 KANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGITQEYVKIHCDSQSAIHLANHQ 366
Query: 1151 IQHSRTKHIDIRHHFIRELVEDGTVTLEHVSTEKQLADIFTK 1192
+ H RTKHIDIR HFIR+++E + +E +++E+ AD+FTK
Sbjct: 367 VYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 112 bits (279), Expect(2) = 4e-26
Identities = 55/113 (48%), Positives = 68/113 (59%)
Frame = +1
Query: 489 TPQQNGVVERKNRTLQESARAMLHAKNLPYHFWAEAINTACYIHNRVTIRQGDTVTQYEL 548
TPQQNGV ER NRTL E RAML + FWAEA+ TACY+ NR D T E+
Sbjct: 85 TPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPMEM 264
Query: 549 WKGKKPTVKYFHVFGSKCYILSDREHRRKLDPKSEVGIFLGYSGNSRAYRVYN 601
WKGK HVFG Y++ + + R KLDPKS IFLGY+ N + Y +++
Sbjct: 265 WKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGYXLWD 423
Score = 25.8 bits (55), Expect(2) = 4e-26
Identities = 9/21 (42%), Positives = 14/21 (65%)
Frame = +3
Query: 465 EFENGQFSEFCSSEGIKHEFS 485
E+ +G+F FC EGI +F+
Sbjct: 12 EYVDGEFLAFCKQEGIVRQFT 74
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 100 bits (248), Expect = 4e-21
Identities = 50/122 (40%), Positives = 75/122 (60%), Gaps = 3/122 (2%)
Frame = +1
Query: 1008 RPDITFAVGVCARYQAEPKTSHLIQVKRIIKYISGTSDYGILYSHNTNS---GLTGYCDA 1064
RPDI ++V V +++ +P+ HLI RI++Y+ GT +YG+L+ + S L Y D+
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 1065 DWAGSADDRKSTSGGCFFLENNLISWFSKKQNCVSLSTAEAEYIAAGSSCTQLMWMKQML 1124
DW G DR+STSG F + ISW +KKQ +LS+ EAEYIA + Q +W+ ++
Sbjct: 301 DWCG---DRRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 1125 KE 1126
KE
Sbjct: 472 KE 477
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 95.5 bits (236), Expect = 1e-19
Identities = 40/112 (35%), Positives = 76/112 (67%)
Frame = +3
Query: 1097 CVSLSTAEAEYIAAGSSCTQLMWMKQMLKEYNVQQDVMTLFCDNLSAINISKNPIQHSRT 1156
C S++ + + +C + +WM+++++E +Q+ +T++CD+ SA++I++NP HSRT
Sbjct: 192 CSSVNDRSGVHGSLPQACKEAIWMQRLMEELGHKQEQITVYCDSQSALHIARNPAFHSRT 371
Query: 1157 KHIDIRHHFIRELVEDGTVTLEHVSTEKQLADIFTKALDATQFEKLRQLLGI 1208
KHI I++HF+RE+VE+G+V ++ + T LAD TK+++ +F R G+
Sbjct: 372 KHIGIQYHFVREVVEEGSVDMQKIHTNDNLADAMTKSINTDKFIWCRSSYGL 527
Score = 73.6 bits (179), Expect = 4e-13
Identities = 38/78 (48%), Positives = 50/78 (63%)
Frame = +1
Query: 1033 VKRIIKYISGTSDYGILYSHNTNSGLTGYCDADWAGSADDRKSTSGGCFFLENNLISWFS 1092
VKRI++YI GTS + + + + GY D+D+AG D RKST+G F L +SW S
Sbjct: 1 VKRIMRYIKGTSGVAVCFG-GSELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLS 177
Query: 1093 KKQNCVSLSTAEAEYIAA 1110
K Q V+LST EAEY+AA
Sbjct: 178 KLQTVVALSTTEAEYMAA 231
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 93.2 bits (230), Expect = 5e-19
Identities = 62/234 (26%), Positives = 108/234 (45%), Gaps = 3/234 (1%)
Frame = -3
Query: 303 SKDNCYMWTSETTFLSARCLMSKEDEVR---IWHQKLGHLNLKSMKRIVAEEAVRGIPKL 359
+K + YM + +C + + +WH +LGH + +++ ++ G+
Sbjct: 677 TKGDLYMLEKLDPVSNYKCSFTSSSSLNKDALWHARLGHPHGRALNLMLP-----GV--- 522
Query: 360 KIQEGKVCGECQIGKQVKMSHQKLQHLTTSKVLELLHMDLMGPMQVESLGGKRYVFVCVD 419
+ E K C C +GK K + + + +L++ DL + S +Y +D
Sbjct: 521 -VFENKNCEACILGKHCKNVFPRTSTVYEN-CFDLIYTDLWTAPSL-SRDNHKYFVTFID 351
Query: 420 DFSRFTWVEFLKEKSDTFEVFKELCLQVQREKGSGIVKIRSDHGKEFENGQFSEFCSSEG 479
+ S++TW+ + K + FK V + I +RSD+G E+ + F G
Sbjct: 350 EKSKYTWLTLIPSKDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHG 171
Query: 480 IKHEFSAPITPQQNGVVERKNRTLQESARAMLHAKNLPYHFWAEAINTACYIHN 533
I H+ S P TPQQNGV +RKN+ L E AR+++ N ++TACY+ N
Sbjct: 170 ILHQTSCPYTPQQNGVAKRKNKHLMEVARSLMFQAN---------VSTACYLIN 36
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 82.4 bits (202), Expect = 1e-15
Identities = 40/95 (42%), Positives = 63/95 (66%)
Frame = +2
Query: 729 FKRNEVWELVPRPDDANVVGTKWIFRNKSDESGNVTRNKSRLVAQGYSQIEGIDFYETFA 788
+ + + +LV +P +G +WI++ K +E G + + K+RLVA+GY + +GIDF E FA
Sbjct: 41 YYQKQTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFA 220
Query: 789 PVARLESIRLLLGVACLLKFRLYQMDVKSAFLNGY 823
PV R+E+I LLL +A ++ +DVK AFLNG+
Sbjct: 221 PVVRIETI*LLLALAATNGC*IHHIDVKIAFLNGH 325
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 59.7 bits (143), Expect(2) = 2e-15
Identities = 32/106 (30%), Positives = 58/106 (54%), Gaps = 1/106 (0%)
Frame = +1
Query: 857 QAPRAWYERLTEFLINNGYDKGGIDKTLFVKKNGG-ELMVAQIYVDDIVFGGMSNQMVEQ 915
Q+PR W++R T + GY + D +F+K + + + +YVDDI G + +++
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 916 FVEQMKSEFEMSLVGELTYFLGLQVKQMEDTLFITQSKYAKGIVKK 961
+ EFE+ +G L YFLG++V + + I+Q KY ++K+
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKE 318
Score = 42.0 bits (97), Expect(2) = 2e-15
Identities = 19/50 (38%), Positives = 31/50 (62%)
Frame = +2
Query: 972 TPAATHIKLTKDEKGTDVDPSLYRSMIGSLLYLTASRPDITFAVGVCARY 1021
TP +KL + GT VD Y+ ++G L+YL+ +RPDI+F V +++
Sbjct: 350 TPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQF 499
>BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440.1 [imported]
- Arabidopsis thaliana, partial (20%)
Length = 731
Score = 74.3 bits (181), Expect = 3e-13
Identities = 39/98 (39%), Positives = 59/98 (59%), Gaps = 1/98 (1%)
Frame = +3
Query: 1112 SSCTQLMWMKQMLKEYNVQQ-DVMTLFCDNLSAINISKNPIQHSRTKHIDIRHHFIRELV 1170
++ Q MW++ +L E + + + + DN S I +++NP+ H R HI R+HFIRE V
Sbjct: 123 TAARQAMWLQDLLSEVTWEPCEEVVIRIDNQSVIALTRNPVFHGRGNHIHKRYHFIRECV 302
Query: 1171 EDGTVTLEHVSTEKQLADIFTKALDATQFEKLRQLLGI 1208
E+G V +EHV EK A I TKAL F ++R +G+
Sbjct: 303 ENGQVEVEHVPGEKHRAYI*TKALGRIIFREIRYYIGM 416
>BE941052 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 480
Score = 72.4 bits (176), Expect = 1e-12
Identities = 34/73 (46%), Positives = 48/73 (65%)
Frame = +2
Query: 1136 LFCDNLSAINISKNPIQHSRTKHIDIRHHFIRELVEDGTVTLEHVSTEKQLADIFTKALD 1195
L CD LSA ++ NP+ HSR KHI I HF+R+LV+ G + ++HV T QLAD TK L
Sbjct: 23 LRCDYLSATYLTHNPVYHSRMKHISIDIHFVRDLVQQGKLKVQHVCTVDQLADCLTKPLS 202
Query: 1196 ATQFEKLRQLLGI 1208
++ + LR +G+
Sbjct: 203 KSRHQLLRNKIGV 241
>BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T15F17.l
{Arabidopsis thaliana}, partial (3%)
Length = 539
Score = 64.7 bits (156), Expect = 2e-10
Identities = 34/104 (32%), Positives = 55/104 (52%)
Frame = -3
Query: 1002 LYLTASRPDITFAVGVCARYQAEPKTSHLIQVKRIIKYISGTSDYGILYSHNTNSGLTGY 1061
+ LT P+ITF++ + +RY + P H +K I KY+ G D G+ YS + + L GY
Sbjct: 531 ILLTLQGPNITFSINLLSRYSSAPTMRH*NGIKHICKYLKGIIDMGLFYSKDCSPDLIGY 352
Query: 1062 CDADWAGSADDRKSTSGGCFFLENNLISWFSKKQNCVSLSTAEA 1105
+A + +S +G F N +ISW S K + ++ S+ A
Sbjct: 351 VNA*YLSDPHKARS*TGYIFTCGNTVISWRSTK*STIATSSNHA 220
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 64.3 bits (155), Expect = 3e-10
Identities = 41/151 (27%), Positives = 69/151 (45%), Gaps = 4/151 (2%)
Frame = -2
Query: 528 ACYIHNRVTIRQGDTVTQYELWKGKKPTVKYFHVFGSKCYILSDREHRRKLDPKSEVGIF 587
ACY+ NR+ R +E+ +KP++ Y VFG CY+L E R KL+ +S +F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 588 LGYSGNSRAYRVYNIRTKVMMESINV-VVDDTSNERTGQAHDEDDLPYE---CTNVEPDE 643
+GYS + Y+ Y+ + ++ S +V +++ D DL + V +
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIEERGYYEEKNQEDLRDLTSDKAGVLRVILEG 345
Query: 644 PAIQFPNEQENTVSQPPLATKEPSIRVQKIH 674
I+ +Q QP ++ EP Q H
Sbjct: 344 LGIKMNQDQSTRSRQPEESSNEPRRAAQTPH 252
>AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (18%)
Length = 567
Score = 63.5 bits (153), Expect = 5e-10
Identities = 33/101 (32%), Positives = 56/101 (54%), Gaps = 3/101 (2%)
Frame = +3
Query: 202 WYFDSGCSRHMTGVKSYLEKMKPHAKSFVTFGDGAKGKIRGIGKL---SDTGSPNLDDVL 258
W DSGC+ HMT + +++ S V +GA ++ GIG + S +G + +VL
Sbjct: 108 WLIDSGCTHHMTHDRDLFKELNKSTISKVRMLNGAHIEVEGIGTVLVKSHSGYKQISNVL 287
Query: 259 LVEGLTANLISISQLCDQGLKVVFKQSGCVVKNKNKDVLMR 299
L +L+S+ QL +G KV+F+ CV+K++N ++R
Sbjct: 288 YAPKLNQSLLSVPQLLTKGYKVLFEHEKCVIKDQNNKEVLR 410
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,521,308
Number of Sequences: 36976
Number of extensions: 508867
Number of successful extensions: 2360
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 2313
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2347
length of query: 1213
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1106
effective length of database: 5,058,295
effective search space: 5594474270
effective search space used: 5594474270
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0172.5


