

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0170.6
(1596 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
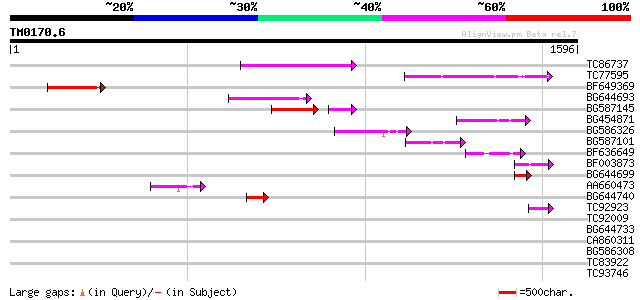
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 249 5e-66
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 172 7e-43
BF649369 152 1e-36
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 144 2e-34
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 107 2e-29
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 121 2e-27
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 86 9e-17
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 65 3e-10
BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativ... 63 8e-10
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 57 4e-08
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 56 1e-07
AA660473 54 5e-07
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 54 6e-07
TC92923 similar to GP|6466937|gb|AAF13073.1| putative retroeleme... 48 3e-05
TC92009 43 0.001
BG644733 weakly similar to GP|15289942|db putative polyprotein {... 42 0.002
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 35 0.23
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 35 0.23
TC83922 similar to GP|15809990|gb|AAL06922.1 AT5g60530/muf9_180 ... 33 0.89
TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical... 33 0.89
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 249 bits (637), Expect = 5e-66
Identities = 130/334 (38%), Positives = 199/334 (58%), Gaps = 8/334 (2%)
Frame = +1
Query: 650 IEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEE 709
++K + ++L G I+ S SA +PV+ V+K CVDYRALN++T D++P+P+I E
Sbjct: 511 LKKTLEDLLDKGFIKASGSAAGAPVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISE 690
Query: 710 LLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQS 769
L + GA +F+KLD+ + +H++R+++ED KTAFRT G +E++V PFGL AP+TFQ
Sbjct: 691 TLRRVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQR 870
Query: 770 LMNEVFRPMLRRGVLVFFDDILVYSK-TWSDHMLHLERVLEVLQHHQLTANKKKCQFATK 828
+N+ L V + DD+L+Y+ + DH + RVL L L+ + KKC+F+
Sbjct: 871 YINKTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVT 1050
Query: 829 FVEYLGHLISE-QGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARP 887
V+Y+G +++ +GV+ DP K+ +++ W P +VKG R FLG YY+ FI Y +I P
Sbjct: 1051TVKYVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEP 1230
Query: 888 LTELTKKD-AFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLY 946
LT LT+KD F WG + + AF LK PVL + D +E D SG +G +L
Sbjct: 1231LTRLTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQ 1410
Query: 947 -----GKHPIAYFSKALGPRNLAKSAYEKELMAV 975
HP+A+ S+ L P ++KEL+AV
Sbjct: 1411EDGTGAAHPVAFHSQRLSPAEYNYPIHDKELLAV 1512
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 172 bits (437), Expect = 7e-43
Identities = 127/436 (29%), Positives = 213/436 (48%), Gaps = 20/436 (4%)
Frame = +2
Query: 1111 SSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLAS 1170
+ L KL++E H + GH G T ++ +W G ++T++ FV CDVC
Sbjct: 161 NELRTKLVQESHDSTAAGHPGRNGTLEIVSRKFFWPGQSQTVRRFVRNCDVCGGIHIWRQ 340
Query: 1171 SPAGLLQPLPIPERVWEDISLNFIIGLP--RSRGFEAIFVVVDRLTKYSHFIPLKHPYTA 1228
+ G L+PLP+P R+ D+S++FI LP R RG + ++V+VDRL+K S + A
Sbjct: 341 AKRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSK-SVTLEEMDTMEA 517
Query: 1229 RSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTE 1288
+ A+ F R HG+P SI+SDR +V FW+E RL G +S++YHP++DG TE
Sbjct: 518 EACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGTE 697
Query: 1289 IVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVL---- 1344
N+ ++ LR + W L + ++S+ TPF +G +
Sbjct: 698 RWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGYHVDPIPTVE 877
Query: 1345 -TRWVL--GETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKR-KHQEFEVGEW 1400
T V+ GE + + K ++D ++ +VAAQ+R A AN +R ++VG+
Sbjct: 878 DTGGVVSEGEAAAQLLVKRMKD---VTGFIQAEIVAAQQRSEASANKRRCPADRYQVGDK 1048
Query: 1401 VFVKIRAHRQVSLANRV----HAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHI 1456
V++ + ++ + ++ H R+ P+ + L +P ++P FH+
Sbjct: 1049 VWLNVSNYKSPRPSKKLDWLHHKYEVTRFVTPHVV---------ELNVP--GTVYPKFHV 1195
Query: 1457 SLLKKAV-----GNYVEEPGLPEQLDGE-ELPSIQPALVLARRDILRQGESISQVLVQWQ 1510
LL++A G V +P P +D + E+ ++ AR + +G Q LV+W+
Sbjct: 1196 DLLRRAASDPLPGQEVVDPQPPPIVDDDGEVEWEVEEILAARWHQVGRGRR-RQALVKWK 1372
Query: 1511 GKTPEEATWEDLATIR 1526
G +ATWE IR
Sbjct: 1373 GFV--DATWEAADAIR 1414
>BF649369
Length = 631
Score = 152 bits (383), Expect = 1e-36
Identities = 77/163 (47%), Positives = 114/163 (69%)
Frame = +3
Query: 106 NEFRQSVKKVELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSL 165
NE R + KKV+LP+F+G D WI+RAEIYF VQ T +++V L++LSMEG TIH++N L
Sbjct: 99 NESRLAGKKVKLPLFEGDDPVAWITRAEIYFDVQNTPDDMRVKLSRLSMEGPTIHWFNLL 278
Query: 166 LATEEELTWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPE 225
+ TE++L+ E+ + AL+ RY G + +E+LS LRQ G VEE++ FE L++Q+ +LPE
Sbjct: 279 METEDDLSREKLKKALIARYDGRRLENPFEELSTLRQIGSVEEFVEAFELLSSQVGRLPE 458
Query: 226 KQYQGYFLHGLKEEIRGKVRSLVAMGGVNRARLLVVTRAVEKE 268
+QY GYF+ GLK IR +VR+L R +++ + + VE E
Sbjct: 459 EQYLGYFMSGLKAHIRRRVRTL---NPTTRMQMMRIAKDVEDE 578
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 144 bits (364), Expect = 2e-34
Identities = 89/238 (37%), Positives = 130/238 (54%), Gaps = 5/238 (2%)
Frame = +2
Query: 616 LPPGRGREHCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVI 675
+PP + I + P+ + YR ++ Q++++L G I+ S V+
Sbjct: 17 VPPEWKIDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVVVL 196
Query: 676 LVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVR 735
+KKKD M +DY LN+V + K+P+P+I+EL D L G+ +F K+DL+ G HQ RV
Sbjct: 197 FLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVI 376
Query: 736 EEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSK 795
EDV KTAFR GHYE +VM FG N P F LMN VF+ L V+VF +DIL+YSK
Sbjct: 377 GEDVPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSK 556
Query: 796 TWSDHMLHLERVLEVLQHHQLTANKKKCQFA-----TKFVEYLGHLISEQGVAVDPNK 848
++H HL L+VL+ L CQ + + + H+IS +G+ VD +
Sbjct: 557 NENEHENHLRLALKVLKDIGL------CQISYV*ILVEVGFFSLHVISGEGLKVDSKR 712
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 107 bits (266), Expect(2) = 2e-29
Identities = 52/133 (39%), Positives = 80/133 (60%)
Frame = +2
Query: 737 EDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKT 796
+D+ KTAF T G Y Y VMPFGL NA ST+Q L+N +F L + V+ DD+LV S
Sbjct: 11 DDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVKSLR 190
Query: 797 WSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWP 856
+DH+ HL+ + L + + N KC F E+LG+++++QG+ V+P ++ ++ P
Sbjct: 191 ATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAILDLP 370
Query: 857 EPKNVKGVRGFLG 869
PKN + V+ G
Sbjct: 371 SPKNSREVQRLTG 409
Score = 42.4 bits (98), Expect(2) = 2e-29
Identities = 26/83 (31%), Positives = 39/83 (46%), Gaps = 4/83 (4%)
Frame = +3
Query: 897 FNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLY----GKHPIA 952
F W ++ ++AFE LK+ LTT PVL+ P+ L S + ++L+ + PI
Sbjct: 495 FVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIAISSTAVSSVLIREDRGEQKPIF 674
Query: 953 YFSKALGPRNLAKSAYEKELMAV 975
Y SK + EK AV
Sbjct: 675 YTSKRMTDPETRYPTLEKMAFAV 743
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 121 bits (304), Expect = 2e-27
Identities = 73/209 (34%), Positives = 108/209 (50%), Gaps = 2/209 (0%)
Frame = +2
Query: 1259 SHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEY 1318
S+FWK++F+L GT MSSAYHP SDGQ+E +N+ E YLRC P W+ WAEY
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 1319 WFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAA 1378
W+NTSY+ + TPF+A+YGR +L R ++ L R+E L QL+ +
Sbjct: 212 WYNTSYNISAAMTPFKALYGRDLSMLIRSKGSSKDTADLQSQLAQREELLSQLQ----SI 379
Query: 1379 QERMRAQANSKR-KHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYP-IIGRVG 1436
R+ + K ++ WV + +L N + +P ++
Sbjct: 380 STRLNKL*SIKLIRNAAILSSSWVSTSL---*NCNLINSLR*HCGNIKSSVHPTLVHY*Q 550
Query: 1437 AVAYRLKLPEGARIHPVFHISLLKKAVGN 1465
+ AY+L LP A++ P+FH+S LK GN
Sbjct: 551 SAAYKLSLPSTAKVPPIFHVSQLKPFHGN 637
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 86.3 bits (212), Expect = 9e-17
Identities = 75/240 (31%), Positives = 113/240 (46%), Gaps = 23/240 (9%)
Frame = +2
Query: 915 TTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMA 974
T+AP+L LP+ + + DAS G+G +L + IAY S+ L ++ E+ A
Sbjct: 8 TSAPILVLPELIT-YVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAA 184
Query: 975 VALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKT 1034
V A++ WR YL G K + TD +SLK Q + Q+ W + Y+ +I Y PGK
Sbjct: 185 VVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPGKA 364
Query: 1035 NQGADALS--RIDEAGE------------LRPLVSYPLWEEQGVIADENQKD----PKLC 1076
N ADALS R+D + E LR V E G+ A NQ D +L
Sbjct: 365 NLVADALSRRRVDVSAEREADDLDGMVRALRLNVLTKATESLGLEA-VNQADLFTRIRLA 541
Query: 1077 Q----IVSEVALDPQAHPGYRVVQ-GTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSG 1131
Q + +VA + + Y+ + GT+L R+ + N SL +++ E H + H G
Sbjct: 542 QGQDENLQKVAQNDRTE--YQTAKDGTILVNGRISVPNDRSLKEEIMSEAHKSRFSVHPG 715
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 64.7 bits (156), Expect = 3e-10
Identities = 46/173 (26%), Positives = 81/173 (46%), Gaps = 4/173 (2%)
Frame = +2
Query: 1114 IPKLLEEFHTTPHGGHSGFLRTYRRIA-TNLYWRGMTKTIQEFV*ACDVCQRHKYLA--- 1169
IP +L H + + GH +T +I +W M K F+ CD CQR ++
Sbjct: 95 IPGILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNIS*RN 274
Query: 1170 SSPAGLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTAR 1229
P + + + + VW ++F+ P S + I V VD ++K+ I A
Sbjct: 275 EMPQNFILEVEVFD-VW---GIDFMGPFPSSYNNKYILVAVDYVSKWVEAIA-SPTNDAT 439
Query: 1230 SVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPE 1282
V ++F I GVP +ISD F++ ++++ + G + K+++AYHP+
Sbjct: 440 VVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQ 598
>BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativa}, partial
(4%)
Length = 653
Score = 63.2 bits (152), Expect = 8e-10
Identities = 49/174 (28%), Positives = 82/174 (46%), Gaps = 4/174 (2%)
Frame = +1
Query: 1282 ESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKP 1341
+SD Q ++N LET+L F ++Q +L WAE +NT++H TPF+ VY
Sbjct: 4 DSDEQAGLLNHTLETHLLYFTSEQQGV*NFFLTWAECLYNTNFHRTAGCTPFKVVY---- 171
Query: 1342 PVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEF----EV 1397
V+ + V +DL R+E L + + +A S+ ++
Sbjct: 172 ------VVAHLQKFVVARDLIYRNEGLHKSST*TSFGRGTRAYEALSRPAYETC*HPRRP 333
Query: 1398 GEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIH 1451
V+ + R + +V K A +GPY +I ++G+VA++L LPE +IH
Sbjct: 334 LSIVYTRDRTYEW-----QVLPKYVA*CYGPYQVIKQIGSVAFKL*LPEQHQIH 480
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 57.4 bits (137), Expect = 4e-08
Identities = 40/117 (34%), Positives = 55/117 (46%), Gaps = 6/117 (5%)
Frame = +2
Query: 1420 KLAARYFGPYPIIGRVGAVAYRLKLPEG-ARIHPVFHISLLKKAVGNYVEEPGLPEQLDG 1478
KL R+ GPY I RVG VAYR+ LP +H VFH+S L+K YV +P Q D
Sbjct: 29 KLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRK----YVPDPSHVIQSDD 196
Query: 1479 EEL-----PSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLATIRSEFP 1530
++ P + R+ +G+ I V V W E TWE + + +P
Sbjct: 197 VQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVESYP 367
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 55.8 bits (133), Expect = 1e-07
Identities = 26/51 (50%), Positives = 36/51 (69%), Gaps = 3/51 (5%)
Frame = +2
Query: 1420 KLAARYFGPYPIIGRVGAVAYRLKLPEG-ARIHPVFHISLLKK--AVGNYV 1467
KL+ RY GP+ +I R+G VAY L LP G + +HPVFH+S+ K+ GNY+
Sbjct: 65 KLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVSMFKRYHGDGNYI 217
>AA660473
Length = 655
Score = 53.9 bits (128), Expect = 5e-07
Identities = 50/173 (28%), Positives = 85/173 (48%), Gaps = 19/173 (10%)
Frame = -2
Query: 397 LAMEVNEDEEETQGELSLMSLCELGM--KTGGI-PRTMKLRGTINGVPVVVLVDSGATHN 453
+ +E+ + E+E + M LCE+ K + P T++L G + GVP+V+LVD GA HN
Sbjct: 651 MTIELKDGEKEDILVYNSMGLCEMTEFNKLKVVKPATLQLVGCLKGVPIVILVDIGANHN 472
Query: 454 FVDCFLVRRLGWE--VVDTPRM-------TVKLGDGYKSQAQGR---CAGL---KIEMGE 498
F V +G + V+ + + +G Y S+ R C G+ ++E+GE
Sbjct: 471 FDFASSVSDVGCKGRVILSQESEVG*WSENINVG*AY*SRGTVRGFHC*GVDAYELELGE 292
Query: 499 YHLRCSPQLFDLGGPDIVLGIEWLKTLGDTIV-NWDTQLMSFWSDKKWITLQG 550
+ D+ LG+ WL+ LG+ +V +WD + + F + + LQG
Sbjct: 291 F--------------DMFLGVAWLEKLGN*VVFDWDERTICFEWKGEVVKLQG 175
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 53.5 bits (127), Expect = 6e-07
Identities = 24/63 (38%), Positives = 39/63 (61%)
Frame = -1
Query: 666 STSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDL 725
S S + ++ V+KKD + MC+DYR N VT +K+P+P I+ L D++ +F +DL
Sbjct: 256 SISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF*NIDL 77
Query: 726 KSG 728
+ G
Sbjct: 76 RLG 68
>TC92923 similar to GP|6466937|gb|AAF13073.1| putative retroelement pol
polyprotein {Arabidopsis thaliana}, partial (1%)
Length = 625
Score = 47.8 bits (112), Expect = 3e-05
Identities = 28/71 (39%), Positives = 42/71 (58%)
Frame = +2
Query: 1460 KKAVGNYVEEPGLPEQLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATW 1519
K +VG+Y E LPE ++ E P +LA R I R+G+ + ++W+ KT E A+
Sbjct: 266 KSSVGSYPVELQLPEGMEVEINDEADPQFMLATRQI-REGDITT**AIKWKAKTLEHASL 442
Query: 1520 EDLATIRSEFP 1530
+D TIRS+FP
Sbjct: 443 KDDFTIRSQFP 475
>TC92009
Length = 974
Score = 42.7 bits (99), Expect = 0.001
Identities = 26/91 (28%), Positives = 44/91 (47%), Gaps = 1/91 (1%)
Frame = +2
Query: 114 KVELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSMEG-GTIHFYNSLLATEEEL 172
K +L F G D AGWI+RAE++F E K+ A +SME + ++ S +
Sbjct: 680 KYKLSQFTGTDPAGWITRAEMFFADNEIHSCDKLQWAFMSMEDEKALLWFYSWCQENPDA 859
Query: 173 TWERFRDALLERYGGNGDGDVYEQLSELRQQ 203
W+ F A++ +G + +Q+ + Q
Sbjct: 860 DWKSFSMAMIREFGARMEHRTDKQMVQNHDQ 952
>BG644733 weakly similar to GP|15289942|db putative polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (1%)
Length = 174
Score = 42.0 bits (97), Expect = 0.002
Identities = 21/43 (48%), Positives = 29/43 (66%)
Frame = -3
Query: 1201 RGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLH 1243
+ +E+I VVVDRLTK + FIP K Y+A+ A I EI+ +H
Sbjct: 130 KSYESI*VVVDRLTKSTLFIPFKTSYSAK*YARILLDEIVCIH 2
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 35.0 bits (79), Expect = 0.23
Identities = 18/44 (40%), Positives = 26/44 (58%)
Frame = +1
Query: 948 KHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKF 991
+HPIAY S+ L + E+E +A AI+++R YL G KF
Sbjct: 55 EHPIAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYLHGPKF 186
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 35.0 bits (79), Expect = 0.23
Identities = 15/52 (28%), Positives = 29/52 (54%)
Frame = -2
Query: 1243 HGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCL 1294
HG+P I++D F+S+ ++E + +S +P+S+GQ E N+ +
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKII 530
>TC83922 similar to GP|15809990|gb|AAL06922.1 AT5g60530/muf9_180
{Arabidopsis thaliana}, partial (21%)
Length = 619
Score = 33.1 bits (74), Expect = 0.89
Identities = 21/91 (23%), Positives = 38/91 (41%)
Frame = +1
Query: 285 GNGSRSGFSGATKGNGTEWIWVKGNKETGQTVNRPNSNPRGDQTKNTGDIRRAGPRDRGF 344
GNG+ +G +G KGNG GN G+ ++ +G + K ++ P++
Sbjct: 109 GNGNGNGNNGNGKGNGNG----NGNNGNGKGNDKEEGKEKGKE-KEKAPKKKKAPKEESS 273
Query: 345 NHLSYPQLMERRQKGLCFKCGGPYHRNHVCP 375
N+ L +++ C + VCP
Sbjct: 274 NYQQLQTLPSGQERAFCKANNTCHFATLVCP 366
>TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical
protein~similar to gag-pol polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (4%)
Length = 1019
Score = 33.1 bits (74), Expect = 0.89
Identities = 14/42 (33%), Positives = 29/42 (68%), Gaps = 1/42 (2%)
Frame = +2
Query: 1180 PIPERVWEDISLNFIIGLPRSRGF-EAIFVVVDRLTKYSHFI 1220
P+P+ WED++++F +GL ++ ++ VV D+ ++ +HFI
Sbjct: 473 PVPKPPWEDVTIDFSLGLL*TQQLKDSKMVVGDKFSRMAHFI 598
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,800,718
Number of Sequences: 36976
Number of extensions: 638339
Number of successful extensions: 3015
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 2950
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3002
length of query: 1596
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1487
effective length of database: 4,984,343
effective search space: 7411718041
effective search space used: 7411718041
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0170.6


