

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0166.9
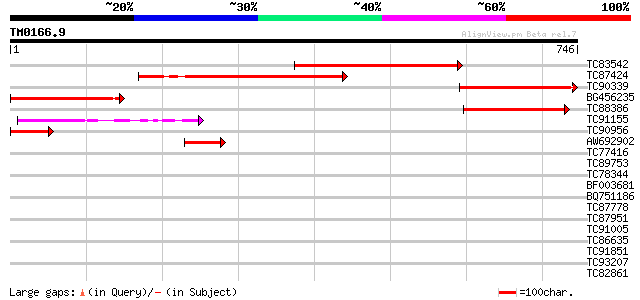
(746 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83542 similar to GP|19387245|gb|AAL87157.1 putative kinesin li... 322 3e-88
TC87424 similar to GP|19699309|gb|AAL91265.1 AT4g10840/F25I24_50... 255 4e-68
TC90339 similar to GP|19387245|gb|AAL87157.1 putative kinesin li... 251 7e-67
BG456235 weakly similar to GP|9802538|gb|A F17L21.29 {Arabidopsi... 211 8e-55
TC88386 similar to GP|19699309|gb|AAL91265.1 AT4g10840/F25I24_50... 189 3e-48
TC91155 similar to GP|9294312|dbj|BAB01483.1 contains similarity... 99 5e-21
TC90956 weakly similar to PIR|F71159|F71159 hypothetical protein... 80 2e-15
AW692902 similar to GP|19699309|gb| AT4g10840/F25I24_50 {Arabido... 48 2e-05
TC77416 similar to GP|18139887|gb|AAL60196.1 O-linked N-acetyl g... 41 0.001
TC89753 similar to GP|14334556|gb|AAK59686.1 unknown protein {Ar... 40 0.003
TC78344 similar to GP|13540405|gb|AAK29456.1 histone H1 {Lens cu... 37 0.028
BF003681 similar to GP|17380952|gb putative kinesin light chain ... 36 0.047
BQ751186 weakly similar to PIR|T40402|T404 forkhead nuclear sign... 34 0.18
TC87778 similar to GP|5103827|gb|AAD39657.1| ESTs gb|F20110 and ... 33 0.31
TC87951 similar to PIR|C86418|C86418 hypothetical protein AAG126... 33 0.31
TC91005 weakly similar to PIR|T00960|T00960 hypothetical protein... 33 0.52
TC86635 weakly similar to GP|9758600|dbj|BAB09233.1 gene_id:MDF2... 32 0.89
TC91851 weakly similar to PIR|F96673|F96673 hypothetical protein... 31 1.5
TC93207 similar to GP|16604571|gb|AAL24087.1 unknown protein {Ar... 30 2.6
TC82861 similar to GP|6630450|gb|AAF19538.1| F23N19.10 {Arabidop... 30 2.6
>TC83542 similar to GP|19387245|gb|AAL87157.1 putative kinesin light chain
gene {Oryza sativa (japonica cultivar-group)}, partial
(30%)
Length = 665
Score = 322 bits (826), Expect = 3e-88
Identities = 157/221 (71%), Positives = 193/221 (87%)
Frame = +1
Query: 375 VAEANVQALQFDEAERLCQMALDIHKANSSAPSLEEAADRRLMGLILDTKGNHEAALEHL 434
+AEA+VQALQFDEA++LCQMALDIHK N S EEAADRRLMGLI D+KG++E+ALEH
Sbjct: 1 LAEAHVQALQFDEAKKLCQMALDIHKGNGLPASFEEAADRRLMGLICDSKGDYESALEHY 180
Query: 435 VLASMAMVANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGS 494
VLAS+ M NGQE++VASVDCSIGD Y S++RYDEA+F+Y+KALTVFK+ KGENHP V S
Sbjct: 181 VLASVVMAENGQELDVASVDCSIGDAYSSLARYDEAIFSYQKALTVFKSTKGENHPTVAS 360
Query: 495 VFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEK 554
VFVRLADL+N+ GK K+SK+YC++ALRI+ PG++ EEIA+GL +V+AIY+SMN+LEK
Sbjct: 361 VFVRLADLYNKIGKFKDSKTYCENALRIFGKKKPGISLEEIANGLIDVAAIYQSMNDLEK 540
Query: 555 ALKLLQKALLIYNDSPGQQSSIAGIEAQMGVMYYMLGNYTE 595
LKLL+KAL IYN+ PGQ S+IAGIEAQMGVMYYMLGNY++
Sbjct: 541 GLKLLKKALKIYNNVPGQHSTIAGIEAQMGVMYYMLGNYSD 663
>TC87424 similar to GP|19699309|gb|AAL91265.1 AT4g10840/F25I24_50
{Arabidopsis thaliana}, partial (32%)
Length = 1014
Score = 255 bits (652), Expect = 4e-68
Identities = 135/278 (48%), Positives = 185/278 (65%), Gaps = 3/278 (1%)
Frame = +2
Query: 170 LDASPKSKPKGKSPPAKAPL-ERKNDKPLLRKQTKGVASGVKSLKSSPLGKSVSLNRVEN 228
L +P + + P ++PL +K P ++K K +P S+
Sbjct: 218 LPETPTQRSESSKTPTQSPLPNKKPPSPSPSSRSK------KKTSETPTTNSLL------ 361
Query: 229 SAESALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLEKLG-NGKPSLELVMCL 287
+E++LD P+ P LLK ARD I+SGD P KAL+ A++A E+ G+PSL+L M L
Sbjct: 362 -SEASLDNPDLGPFLLKVARDTIASGDGPAKALDYAIRASKSFERCAVEGEPSLDLAMSL 538
Query: 288 HVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAMLGQLENSI 347
HV AAIYC+L ++ EA+P+LER+I +P + HALA F+G+MQLGDT++MLGQ++ SI
Sbjct: 539 HVLAAIYCSLNRFEEAVPVLERAIHVPDVERGADHALAAFSGYMQLGDTFSMLGQVDKSI 718
Query: 348 MCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSAPS 407
+ Y G ++Q Q LGETDPRVGETCRY+AEANVQA+QFD+AE LC L+IH+A+S S
Sbjct: 719 LMYDQGLQIQIQTLGETDPRVGETCRYLAEANVQAMQFDKAEELCNKTLEIHRAHSEPAS 898
Query: 408 LEEAADRRLMGLILDTKGNHEAALEHLVLASM-AMVAN 444
LEEAADRRLM LI + KG++E+ALEHL + M+AN
Sbjct: 899 LEEAADRRLMALICEAKGDYESALEHLCSCRVWQMIAN 1012
>TC90339 similar to GP|19387245|gb|AAL87157.1 putative kinesin light chain
gene {Oryza sativa (japonica cultivar-group)}, partial
(19%)
Length = 779
Score = 251 bits (641), Expect = 7e-67
Identities = 131/155 (84%), Positives = 138/155 (88%)
Frame = +2
Query: 592 NYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILEH 651
NYTESYNTLK AI KLRAIGEKKSSF GIALNQ+GLACVQ YA EA ELFEEAKS+LEH
Sbjct: 2 NYTESYNTLKKAIAKLRAIGEKKSSFFGIALNQLGLACVQRYALREAIELFEEAKSVLEH 181
Query: 652 EYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLSE 711
E GPYHPETLGVYSNLAGTYDAIGRLD+AI LE+VVS+REEKLGTANPDVDDEKRRLSE
Sbjct: 182 ELGPYHPETLGVYSNLAGTYDAIGRLDDAILTLEHVVSMREEKLGTANPDVDDEKRRLSE 361
Query: 712 LLKEAGRVRSRKVMSLENLLDANARIPNINLVIKA 746
LLKEAG+VRSRKV SLENL D +A N NLVIKA
Sbjct: 362 LLKEAGKVRSRKVRSLENLFDGDAHTLN-NLVIKA 463
>BG456235 weakly similar to GP|9802538|gb|A F17L21.29 {Arabidopsis thaliana},
partial (7%)
Length = 538
Score = 211 bits (537), Expect = 8e-55
Identities = 112/152 (73%), Positives = 124/152 (80%), Gaps = 1/152 (0%)
Frame = +3
Query: 1 MPGIVTNGVCD-GAVNEMNGNHSPSKETLAPVKSPRGSLSPQRGQNGGPNFPVDGVIEPS 59
MPGIVTNGV D G VNEMNGN SP+K +L P+KSPR S+SPQR Q+ GPNF DGV+EPS
Sbjct: 75 MPGIVTNGVHDDGEVNEMNGNRSPTKASLTPIKSPRSSMSPQRRQSVGPNFQDDGVVEPS 254
Query: 60 IEQLYENVCDMQSSDQSPSRKSFGSDGDESRIDSELRQLVGGRMREVEIMEEEVEVEVEK 119
IEQLYENVCDMQSSDQSPSR+SFGSDGDESRIDSEL LVGGRMRE+EIMEEEVEVEV
Sbjct: 255 IEQLYENVCDMQSSDQSPSRQSFGSDGDESRIDSELHHLVGGRMRELEIMEEEVEVEVGX 434
Query: 120 ERGGSSSGEISSGVGGLSSNEKKLDKVHEIQS 151
SSSGE SSG+G L S++ +D IQS
Sbjct: 435 XPXXSSSGETSSGMGNL-SDDXXMDMXAXIQS 527
>TC88386 similar to GP|19699309|gb|AAL91265.1 AT4g10840/F25I24_50
{Arabidopsis thaliana}, partial (21%)
Length = 736
Score = 189 bits (480), Expect = 3e-48
Identities = 91/139 (65%), Positives = 113/139 (80%)
Frame = +2
Query: 598 NTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYH 657
+ ++A+ KLR GE+KS+F G+ LNQMGLACVQL+ EA ELFEEA+ ILE E GP H
Sbjct: 8 SAFESAVLKLRTSGERKSAFFGVVLNQMGLACVQLFKIDEAAELFEEARGILEQECGPCH 187
Query: 658 PETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLSELLKEAG 717
+TLGVYSNLA TYDA+GR+ +AI+ILEYV+ +REEKLG ANPD +DEKRRL+ELLKEAG
Sbjct: 188 QDTLGVYSNLAATYDAMGRVGDAIEILEYVLKLREEKLGIANPDFEDEKRRLAELLKEAG 367
Query: 718 RVRSRKVMSLENLLDANAR 736
+ R RK SLENL+D N++
Sbjct: 368 KTRDRKAKSLENLIDPNSK 424
>TC91155 similar to GP|9294312|dbj|BAB01483.1 contains similarity to kinesin
light chain~gene_id:K24A2.5 {Arabidopsis thaliana},
partial (8%)
Length = 746
Score = 99.4 bits (246), Expect = 5e-21
Identities = 79/246 (32%), Positives = 120/246 (48%), Gaps = 1/246 (0%)
Frame = +1
Query: 11 DGAVNEMNGNHSPSKETL-APVKSPRGSLSPQRGQNGGPNFPVDGVIEPSIEQLYENVCD 69
+ A E + +++P K+ A SPR +LSP+ Q+ + +DGV++ SIEQLY NV +
Sbjct: 157 NNATEEASESYTPHKDNFTAQQASPRSTLSPRSIQSDSIDLAIDGVVDTSIEQLYTNVYE 336
Query: 70 MQSSDQSPSRKSFGSDGDESRIDSELRQLVGGRMREVEIMEEEVEVEVEKERGGSSSGEI 129
M+SSDQSPSR S S G+ESRIDSEL LV G + ++EI +E V E
Sbjct: 337 MRSSDQSPSRASLYSYGEESRIDSELGHLV-GYIGDLEIRKEIVTTE------------- 474
Query: 130 SSGVGGLSSNEKKLDKVHEIQSATTSSVSTEKSVKALNSQLDASPKSKPKGKSPPAKAPL 189
+ E +D + ++ ++++ + + S KS K +S L
Sbjct: 475 -------NKEEPNVDAIEKVIVSSSTRIG------------EGSSKSAAKSRS------L 579
Query: 190 ERKNDKPLLRKQTKGVASGVKSLKSSPLGKSVSLNRVENSAESALDKPERAPILLKQARD 249
++N+K KG K LG +++ LD P+ P LLK RD
Sbjct: 580 HKRNEK----GTRKGNGFYNMMRKHKRLG-------LKDGIVDELDNPDLGPFLLKNTRD 726
Query: 250 LISSGD 255
+ISSG+
Sbjct: 727 MISSGE 744
>TC90956 weakly similar to PIR|F71159|F71159 hypothetical protein PH0475 -
Pyrococcus horikoshii, partial (15%)
Length = 606
Score = 80.5 bits (197), Expect = 2e-15
Identities = 40/58 (68%), Positives = 46/58 (78%), Gaps = 1/58 (1%)
Frame = +3
Query: 1 MPGIVTNGVCD-GAVNEMNGNHSPSKETLAPVKSPRGSLSPQRGQNGGPNFPVDGVIE 57
MPGIVTNGV D G VNEMNGN SP+K +L P+KSPR S+SPQR + GPNF DGV+E
Sbjct: 432 MPGIVTNGVHDDGEVNEMNGNRSPTKASLTPIKSPRSSMSPQRR*SVGPNFQDDGVVE 605
>AW692902 similar to GP|19699309|gb| AT4g10840/F25I24_50 {Arabidopsis
thaliana}, partial (5%)
Length = 601
Score = 47.8 bits (112), Expect = 2e-05
Identities = 25/55 (45%), Positives = 36/55 (65%), Gaps = 1/55 (1%)
Frame = +3
Query: 231 ESALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLL-EKLGNGKPSLELV 284
ES+L+ P+ P LLK ARD I+SG+NP+KAL+ A+ + + G+G SL V
Sbjct: 432 ESSLENPDLGPFLLKMARDTIASGENPEKALDYAIPGIQVF*TGFGSGCGSLPRV 596
>TC77416 similar to GP|18139887|gb|AAL60196.1 O-linked N-acetyl glucosamine
transferase {Arabidopsis thaliana}, partial (94%)
Length = 3465
Score = 41.2 bits (95), Expect = 0.001
Identities = 51/266 (19%), Positives = 102/266 (38%)
Frame = +2
Query: 456 SIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSY 515
++G+ Y ++ EA+ Y+ AL + P G + LA +H G++
Sbjct: 1016 NLGNVYKALGMPQEAIACYQHAL--------QTRPNYGMAYGNLASIHYEQGQL------ 1153
Query: 516 CDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSS 575
D A+ Y+ + +P + + N+ + + +E+A++ + L + + P ++
Sbjct: 1154 -DMAILHYKQAI-ACDPRFLEA-YNNLGNALKDVGRVEEAIQCYNQCLSLQPNHPQALTN 1324
Query: 576 IAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAF 635
+ I + + M+ Y N T L A N + + Q +
Sbjct: 1325 LGNIYME----WNMVAAAASYYKATLNVTTGLSA-----------PYNNLAIIYKQQGNY 1459
Query: 636 GEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKL 695
+A + E I P N TY IGR+ +AIQ +Y+ ++
Sbjct: 1460 ADAISCYNEVLRI--------DPLAADGLVNRGNTYKEIGRVSDAIQ--DYIRAI----- 1594
Query: 696 GTANPDVDDEKRRLSELLKEAGRVRS 721
T P + + L+ K++G V +
Sbjct: 1595 -TVRPTMAEAHANLASAYKDSGHVEA 1669
Score = 39.7 bits (91), Expect = 0.004
Identities = 49/224 (21%), Positives = 90/224 (39%), Gaps = 19/224 (8%)
Frame = +2
Query: 487 ENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIY 546
E P + LA + R G++ E+ C AL I NP+ + +
Sbjct: 677 ELRPNFADAWSNLASAYMRKGRLTEAAQCCRQALAI--NPL--------------MVDAH 808
Query: 547 ESMNELEKALKLLQKALLIYNDSPGQQSSIAGIEAQMGVMYYMLGNYTESYNTLKNAITK 606
++ L KA L+Q+A Y ++ Q + A + + ++ G++ + K A+ K
Sbjct: 809 SNLGNLMKAQGLVQEAYSCYLEALRIQPTFAIAWSNLAGLFMESGDFNRALQYYKEAV-K 985
Query: 607 LR-----AIGEKKSSFIGIALNQMGLACVQL---------YAFGEATELFEEA----KSI 648
L+ A + + + + Q +AC Q A+G + E +I
Sbjct: 986 LKPSFPDAYLNLGNVYKALGMPQEAIACYQHALQTRPNYGMAYGNLASIHYEQGQLDMAI 1165
Query: 649 LEHEYG-PYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVR 691
L ++ P L Y+NL +GR++EAIQ +S++
Sbjct: 1166LHYKQAIACDPRFLEAYNNLGNALKDVGRVEEAIQCYNQCLSLQ 1297
>TC89753 similar to GP|14334556|gb|AAK59686.1 unknown protein {Arabidopsis
thaliana}, partial (6%)
Length = 1062
Score = 40.4 bits (93), Expect = 0.003
Identities = 31/111 (27%), Positives = 53/111 (46%), Gaps = 5/111 (4%)
Frame = +1
Query: 144 DKVHEIQSATTSSVSTEKSVKALNSQLDASPKSKPKGKSPPAKAPLE--RKNDKPLLRKQ 201
+ +++++S T + S K ++ N +D S + K K K E R P+ K
Sbjct: 361 ENINQLESTATGN-SAMKEIEGSNDNVDGSNLTVSKEKEVKIKVSTEQSRAQKGPVKNKN 537
Query: 202 TK-GVASGVKS--LKSSPLGKSVSLNRVENSAESALDKPERAPILLKQARD 249
K G +SGV + +K+S +GK + ++ SALD R PI + + D
Sbjct: 538 AKVGSSSGVNASLVKNSKIGKDKQASPAVSNGTSALDSRPRQPIKNRSSND 690
>TC78344 similar to GP|13540405|gb|AAK29456.1 histone H1 {Lens culinaris},
partial (86%)
Length = 1215
Score = 37.0 bits (84), Expect = 0.028
Identities = 25/97 (25%), Positives = 38/97 (38%), Gaps = 12/97 (12%)
Frame = +2
Query: 161 KSVKALNSQLDASPKSKPKGKSPPAKAPLERKNDKPLLRK------------QTKGVASG 208
KS A PK KPK K+P AKAP + K K + K A
Sbjct: 470 KSAAPAKKPAAAKPKPKPKAKAPVAKAPAAKSKAKAAPAKAKAKAKAKAAPAKAKPAAKA 649
Query: 209 VKSLKSSPLGKSVSLNRVENSAESALDKPERAPILLK 245
+ K+ P K+ + + + A++ + K P+ K
Sbjct: 650 KPAAKAKPAAKAKPVAKAKPKAKAPVAKANAVPVAAK 760
>BF003681 similar to GP|17380952|gb putative kinesin light chain {Arabidopsis
thaliana}, partial (23%)
Length = 462
Score = 36.2 bits (82), Expect = 0.047
Identities = 32/155 (20%), Positives = 66/155 (41%), Gaps = 7/155 (4%)
Frame = +2
Query: 231 ESALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLEKLGNGKPSLELVMCLHVT 290
E + ++ E LK L G++P+KALE A +A+ + E K S M L +
Sbjct: 5 EISFNEKELGLAALKIGLKLDREGEDPEKALEFANRALKVFE--NEKKDSFPYAMSLQLM 178
Query: 291 AAIYCNLGQYNEAIPILERSIEIPVIGESQQHAL-------AKFAGHMQLGDTYAMLGQL 343
++ L ++++++ L ++ V+G + + A +L + +G+
Sbjct: 179 GSLNYGLNRFSDSLGYLNKANR--VLGRLENEGVCVDDIRPVLHAVQFELANVKTAMGRR 352
Query: 344 ENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEA 378
E ++ E++ E +G+ +AEA
Sbjct: 353 EEALENLRKCLEIKEMTFDEDSEELGKGNMDLAEA 457
>BQ751186 weakly similar to PIR|T40402|T404 forkhead nuclear signaling
protein - fission yeast (Schizosaccharomyces pombe),
partial (19%)
Length = 711
Score = 34.3 bits (77), Expect = 0.18
Identities = 29/101 (28%), Positives = 47/101 (45%), Gaps = 17/101 (16%)
Frame = +2
Query: 133 VGGLSSNEKKLDKVHEIQS-ATTSSVSTEKSVKALNSQLDASPKS--------------- 176
V G+ K L + + S ++TSS S+ +V NSQ + +P S
Sbjct: 8 VRGMFMPRKTLQRSNSSSSVSSTSSNSSTTTVATNNSQPNGTPASTNSDMSSWSAASRKR 187
Query: 177 -KPKGKSPPAKAPLERKNDKPLLRKQTKGVASGVKSLKSSP 216
PKG P K + + +P++R Q G+ +GV SL+ +P
Sbjct: 188 PPPKGPWPAGKPDGQAEFGRPIMRPQMNGI-NGVTSLQPAP 307
>TC87778 similar to GP|5103827|gb|AAD39657.1| ESTs gb|F20110 and gb|F20109
come from this gene. {Arabidopsis thaliana}, partial
(24%)
Length = 1350
Score = 33.5 bits (75), Expect = 0.31
Identities = 30/128 (23%), Positives = 53/128 (40%)
Frame = +2
Query: 436 LASMAMVANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSV 495
LA M V AS + +++A +KAL + + G +HP
Sbjct: 557 LAKMMTVCGPYHRNTASAYSLLAVVLYHTGDFNQATIYQQKALDINERELGLDHPDTMKS 736
Query: 496 FVRLADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKA 555
+ L+ + R I+ + Y + AL + G++ A+ NV+ + E M + A
Sbjct: 737 YGDLSVFYYRLQHIELALKYVNRALFLLHFTC-GLSHPNTAATYINVAMMEEGMGNVHVA 913
Query: 556 LKLLQKAL 563
L+ L +AL
Sbjct: 914 LRYLHEAL 937
>TC87951 similar to PIR|C86418|C86418 hypothetical protein AAG12629.1
[imported] - Arabidopsis thaliana, partial (13%)
Length = 922
Score = 33.5 bits (75), Expect = 0.31
Identities = 41/171 (23%), Positives = 65/171 (37%), Gaps = 5/171 (2%)
Frame = +3
Query: 24 SKETLAPVKSPRGSLSPQRGQNGGPNFPVD-GVIEPSIEQLYENVCDMQSSDQSPSRKSF 82
S+E+L + SP+ R NGG N ++ G + S ++ Y N S D + S
Sbjct: 381 SEESLEKIDSPKDLNYGSRKNNGGSNDVIETGTAKNSKDESYNNSVAEVSDDLVKAVTSV 560
Query: 83 GSDGDESRIDSELRQLVGGRMREVE--IMEEEVEVEVEKERGGSSSGEISSGVGGLSSNE 140
++ L + +G R E + + E +V V + GG S
Sbjct: 561 TEMQSGGTVNDILEKPLGSRAAETDLAVKRNEDKVHVLPDEGGRIS-------------S 701
Query: 141 KKLDKVHEIQSATTSSVSTEKSVKALN--SQLDASPKSKPKGKSPPAKAPL 189
K + E +SSVS +A N + S ++ PP APL
Sbjct: 702 LKEPETREFDGKVSSSVSQSSIPEATNDVENVKGSNAAESSENQPPV-APL 851
>TC91005 weakly similar to PIR|T00960|T00960 hypothetical protein F20D22.10
- Arabidopsis thaliana, partial (36%)
Length = 600
Score = 32.7 bits (73), Expect = 0.52
Identities = 39/197 (19%), Positives = 78/197 (38%), Gaps = 11/197 (5%)
Frame = +1
Query: 250 LISSGDNPQKALELALQAMNLLEKLG-NGKPSLELVMCLHVTAAI--------YCNLGQ- 299
++S +K +AL E L N K LE + + ++A+ Y +G+
Sbjct: 7 VLSQSKKKRKRATMALWMEKGSEPLTENEKADLEAIAAIKESSALELKEKGNQYVKMGKK 186
Query: 300 -YNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAMLGQLENSIMCYSSGFEVQR 358
Y++AI R+I+ + +S+ L H+ L +LG ++ + ++
Sbjct: 187 HYSDAIDCYTRAIDQKALTDSETSVLFSNRAHVNL-----LLGNFRRALNDANDSIKLS- 348
Query: 359 QVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSAPSLEEAADRRLMG 418
P + A+A+ DEA+ CQM L + N L++ + +++
Sbjct: 349 -------PSNIKAIYRAAKASFSLNLLDEAQDYCQMGLHLDPKNEELKRLDKQIETKIL- 504
Query: 419 LILDTKGNHEAALEHLV 435
HEA + + +
Sbjct: 505 ----ENEKHEAEVSNAI 543
>TC86635 weakly similar to GP|9758600|dbj|BAB09233.1
gene_id:MDF20.10~pir||T08929~similar to unknown protein
{Arabidopsis thaliana}, partial (32%)
Length = 1963
Score = 32.0 bits (71), Expect = 0.89
Identities = 29/121 (23%), Positives = 48/121 (38%), Gaps = 3/121 (2%)
Frame = +2
Query: 79 RKSFGSDGDESRIDSELRQLVGGRMREVEIMEE---EVEVEVEKERGGSSSGEISSGVGG 135
+K+ S G E + ++ G + E +E ++ + E E S E S G
Sbjct: 848 KKNEDSSGVEKKSTTDTEDESEGEEKNEENDDEPENDIPEKSEDETPQKSEREDKSDSGS 1027
Query: 136 LSSNEKKLDKVHEIQSATTSSVSTEKSVKALNSQLDASPKSKPKGKSPPAKAPLERKNDK 195
S + KK + + SA S K+ K+ + SP PP +AP + N +
Sbjct: 1028ESEDVKKRKRPSKTSSAKKESAGRSKTEKSTVTNKSRSP--------PPKRAPKKSSNTR 1183
Query: 196 P 196
P
Sbjct: 1184P 1186
>TC91851 weakly similar to PIR|F96673|F96673 hypothetical protein F13O11.30
[imported] - Arabidopsis thaliana, partial (12%)
Length = 981
Score = 31.2 bits (69), Expect = 1.5
Identities = 26/101 (25%), Positives = 45/101 (43%), Gaps = 6/101 (5%)
Frame = +3
Query: 155 SSVSTEKSVKALNSQLDASPKSKPKGKSPPAK-AP-----LERKNDKPLLRKQTKGVASG 208
S +S EKS +++NS+ KS +PP K AP E +N + ++ K
Sbjct: 240 SRLSVEKSPRSVNSKPAVERKSAKATATPPDKQAPRAAKGSELQNQLNVAQEDLKKAKEQ 419
Query: 209 VKSLKSSPLGKSVSLNRVENSAESALDKPERAPILLKQARD 249
+ + + L + AE A +K + A + KQA++
Sbjct: 420 ILQAEKEKVKAIDELKEAQRVAEEANEKLQEALVAQKQAKE 542
>TC93207 similar to GP|16604571|gb|AAL24087.1 unknown protein {Arabidopsis
thaliana}, partial (28%)
Length = 742
Score = 30.4 bits (67), Expect = 2.6
Identities = 17/48 (35%), Positives = 27/48 (55%)
Frame = -3
Query: 157 VSTEKSVKALNSQLDASPKSKPKGKSPPAKAPLERKNDKPLLRKQTKG 204
V T+ K + ++ + + + PKG P +KA + +K K LL K TKG
Sbjct: 653 VGTQI*DKCVQTRSETN*RCNPKGHPPESKAEIFKKLLKMLLMKATKG 510
>TC82861 similar to GP|6630450|gb|AAF19538.1| F23N19.10 {Arabidopsis
thaliana}, partial (29%)
Length = 750
Score = 30.4 bits (67), Expect = 2.6
Identities = 16/48 (33%), Positives = 26/48 (53%), Gaps = 6/48 (12%)
Frame = +2
Query: 457 IGDTYLSMSRYDEAVFAYEKALTV------FKTGKGENHPAVGSVFVR 498
+G +L +S+YD+AV AY+K L + K+G + A + F R
Sbjct: 308 LGAAHLGLSQYDDAVSAYKKGLEIDPNNEPLKSGLADAQKAAAASFSR 451
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.311 0.130 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,570,572
Number of Sequences: 36976
Number of extensions: 207809
Number of successful extensions: 926
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 904
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 920
length of query: 746
length of database: 9,014,727
effective HSP length: 103
effective length of query: 643
effective length of database: 5,206,199
effective search space: 3347585957
effective search space used: 3347585957
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0166.9


