

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0166.5
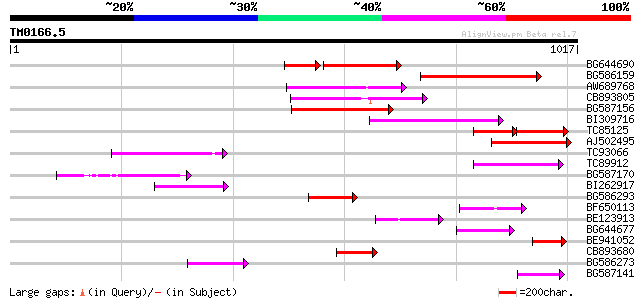
(1017 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 199 3e-68
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 201 9e-52
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 170 3e-42
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 167 2e-41
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 164 2e-40
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 150 2e-36
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 84 5e-33
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 138 1e-32
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 132 9e-31
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 117 2e-26
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 109 6e-24
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 105 9e-23
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 90 4e-18
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 90 5e-18
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 71 2e-12
BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T... 69 1e-11
BE941052 weakly similar to PIR|B85188|B85 retrotransposon like p... 64 2e-10
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 64 3e-10
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 64 3e-10
BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440... 58 2e-08
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 199 bits (507), Expect(2) = 3e-68
Identities = 94/141 (66%), Positives = 119/141 (83%)
Frame = -2
Query: 563 VTRNKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNG 622
+TRNK++LV QGY+Q+EGIDY E FSPVAR+EAIR+LI+F+ L+QMDVKSAF+NG
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 623 YISEEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDN 682
+ EEV+VKQPPGFED + P+HV++L K+LYGLKQAPRAWYERLS FLL+N F RGK DN
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 683 TLFCITYKNDILIVQIYVDDI 703
TLF + + ++LI+Q+YVDDI
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 79.0 bits (193), Expect(2) = 3e-68
Identities = 37/64 (57%), Positives = 46/64 (71%)
Frame = -3
Query: 494 LLSLKGLVSLIEPKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVF 553
L+S +S IEPK+V EAL D WI MQEEL QF ++ VW L+ +P+G VIGT+WVF
Sbjct: 621 LVSFSAFISSIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVF 442
Query: 554 RNKL 557
RNKL
Sbjct: 441 RNKL 430
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 201 bits (512), Expect = 9e-52
Identities = 95/217 (43%), Positives = 142/217 (64%), Gaps = 1/217 (0%)
Frame = +1
Query: 738 IQIDQRPGVTYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGM 797
+++ Q YI Q+KY +LL++F M N+S+ P+ P C L K+E KV Y+ +
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 798 IGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYT 857
+G L+YL A+RPD+++ + L +RF + P E H+ AVKR+L+YL GT NLG+MY++
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 858 LSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQT 917
L + D+D+AGD +RKSTSG L S V+WSSK+Q + LST +AE+++AA C Q+
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 918 IWMKNHLEDYGLSLK-KVPIYCDNTAAISLSKNPILH 953
+WM+ LE G + + +YCDN + I LSKNP+LH
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 170 bits (430), Expect = 3e-42
Identities = 95/218 (43%), Positives = 126/218 (57%), Gaps = 3/218 (1%)
Frame = +1
Query: 497 LKGLVSLIEPKSVD---EALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVF 553
LK V L EP D L D W+ M+ E N W L+ P IG KWV+
Sbjct: 25 LKPKVFLTEPCPQDCENLPLSDPRWLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVY 204
Query: 554 RNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQM 613
R K N G V + KARLVA+G+SQ G DYTETFSPV + IRL+++ ++ + + Q+
Sbjct: 205 RVKENPDGSVNKFKARLVAKGFSQTLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQI 384
Query: 614 DVKSAFLNGYISEEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQN 673
D+ +AFLNG++ EEVY+ QP GFE V KL KSLYGLKQAPRAWYE L+S +Q
Sbjct: 385 DINNAFLNGFLQEEVYMSQPQGFEAAN-KSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQF 561
Query: 674 EFVRGKGDNTLFCITYKNDILIVQIYVDDIIFGSANPS 711
F + + D +L + + IYVDDI+ ++ S
Sbjct: 562 GFTKSRCDPSLLIYNQNGACIYLXIYVDDILITGSSAS 675
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 167 bits (422), Expect = 2e-41
Identities = 90/257 (35%), Positives = 148/257 (57%), Gaps = 12/257 (4%)
Frame = +3
Query: 505 EPKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVT 564
+P + +EA++ + W M E++ +N+ W L G IG KW+F+ KLNE GE+
Sbjct: 39 DPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNENGEIE 218
Query: 565 RNKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSV---NHNITLHQMDVKSAFLN 621
+ KARLVA+GYSQQ G+DYTE F+PVAR + IR++I+ + + + M + +
Sbjct: 219 KYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M*KAHSCME 398
Query: 622 GYISEEVYVKQPPGFEDDKYPDHVY--------KLKKSLYGLKQAPRAWYERLSSFLLQN 673
+ + + + +H ++K++LYGLKQAPRAWY R+ ++ +
Sbjct: 399 N*MRKFLLI------------NHRVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKE 542
Query: 674 EFVRGKGDNTLFC-ITYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGEL 732
F + ++TLF ++ ILI+ +YVDD+IF + ++ +EF K M+ EF MS +G++
Sbjct: 543 GFEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKM 722
Query: 733 KYFLGIQIDQRPGVTYI 749
YFLG+++ Q YI
Sbjct: 723 HYFLGVEVTQNEKGIYI 773
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 164 bits (414), Expect = 2e-40
Identities = 82/183 (44%), Positives = 115/183 (62%)
Frame = -1
Query: 506 PKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTR 565
P+S +EA+EDK W + E KND W PKG + ++W+F K G + R
Sbjct: 558 PRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYKADGSIER 379
Query: 566 NKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYIS 625
K RLVA+G++ G DY ETF+PVA+L IR+++S +VN L QMDVK+AFL G +
Sbjct: 378 KKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQGELE 199
Query: 626 EEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLF 685
+EVY+ PPG E +V +LKK++YGLKQ+PRAWY +LS+ L F + + D+TLF
Sbjct: 198 DEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELDHTLF 19
Query: 686 CIT 688
+T
Sbjct: 18 TLT 10
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 150 bits (379), Expect = 2e-36
Identities = 86/241 (35%), Positives = 131/241 (53%)
Frame = +2
Query: 645 VYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITYKNDILIVQIYVDDII 704
V +L+KS+YGLKQA R WY +LS L+ +++ D +LF + + +YVDDI+
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDIV 199
Query: 705 FGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYTLELLKKFNM 764
+ S + + F++ +G L+YFLG+++ + ++Q+KYTLELL+
Sbjct: 200 LAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSGN 379
Query: 765 SDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSD 824
+ TP + L + + YR +IG L+YLT +RPDI F+V ++F S
Sbjct: 380 LAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVSK 559
Query: 825 PRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLG 884
P++ H A R+L+YLK GL Y TS LS F D+D+A RKS +G FLG
Sbjct: 560 PQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFLG 739
Query: 885 S 885
S
Sbjct: 740 S 742
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 84.3 bits (207), Expect(2) = 5e-33
Identities = 31/93 (33%), Positives = 62/93 (66%)
Frame = +3
Query: 909 SAATCCTQTIWMKNHLEDYGLSLKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRD 968
S C + IWM+ +E+ G +++ +YCD+ +A+ +++NP HSR KHI ++YH++R+
Sbjct: 228 SLPQACKEAIWMQRLMEELGHKQEQITVYCDSQSALHIARNPAFHSRTKHIGIQYHFVRE 407
Query: 969 HVQKGTLSLEYVDTDHQWADIFTKPLAEDRFLF 1001
V++G++ ++ + T+ AD TK + D+F++
Sbjct: 408 VVEEGSVDMQKIHTNDNLADAMTKSINTDKFIW 506
Score = 76.3 bits (186), Expect(2) = 5e-33
Identities = 38/78 (48%), Positives = 54/78 (68%)
Frame = +1
Query: 833 VKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSS 892
VKRI++Y+KGT+ + + + SE T+ G+ D+DFAGD +RKST+G L V+W S
Sbjct: 1 VKRIMRYIKGTSGVAVCFGG-SELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLS 177
Query: 893 KRQNTIALSTAEAEYVSA 910
K Q +ALST EAEY++A
Sbjct: 178 KLQTVVALSTTEAEYMAA 231
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 138 bits (347), Expect = 1e-32
Identities = 66/146 (45%), Positives = 99/146 (67%), Gaps = 1/146 (0%)
Frame = +2
Query: 864 ADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNH 923
+D+AGD RKSTSG LG+ ++WSSK+Q +A STAEAEY+++ +C TQT+W++
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 924 LED-YGLSLKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDT 982
LE + IYCDN +AI+LSKNP+ H R+KHI++++H IR+ + + + +EY T
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 983 DHQWADIFTKPLAEDRFLFILENLNM 1008
+ + ADIFTKPL + F + + L M
Sbjct: 362 EEKIADIFTKPLKIESFYKLKKMLGM 439
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 132 bits (331), Expect = 9e-31
Identities = 77/211 (36%), Positives = 115/211 (54%), Gaps = 2/211 (0%)
Frame = +1
Query: 183 LELLHIDLFGPVKTESIGGKKYGLVIVDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEF 242
L+ +H DL+GP K S GG++Y + I+DD+ R WV FLR+K+ET F + V+ +
Sbjct: 97 LDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQT 276
Query: 243 QTSVITVRSDHGGEFENKAFEELFNSQGISHNFSCPRTPQQNGVVERKNRTLQEMARTMM 302
+V + +D+ EF + F E + GI+ + + PR PQQNGV ER RTL E AR M+
Sbjct: 277 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 456
Query: 303 QESSM--AKHLWAEAINTAYYIHNIISIRPILEKTPYELCKGRQPDISYFHPFGSTCYML 360
+ + + LW EA +TA ++ N + K P ++ G D S FG Y L
Sbjct: 457 SNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYAL 636
Query: 361 NTKEQLGKFDSKALKCYFLGYSERSKGFRIY 391
GK +A +C FL Y+ SKG+R++
Sbjct: 637 VND---GKLAPRAGECIFLSYASESKGYRLW 720
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 117 bits (294), Expect = 2e-26
Identities = 57/163 (34%), Positives = 97/163 (58%), Gaps = 2/163 (1%)
Frame = +1
Query: 832 AVKRILKYLKGTTNLGLMYRKTS--EYTLSGFCDADFAGDRVERKSTSGSCHFLGSNLVT 889
A+K +LKYL + L Y K + E L G+ DAD+AG+ RKS SG L ++
Sbjct: 4 ALKWVLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTIS 183
Query: 890 WSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGLSLKKVPIYCDNTAAISLSKN 949
W + +Q+ + LST +AEY++ IW+K + + G++ + V I+CD+ +AI L+ +
Sbjct: 184 WKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGITQEYVKIHCDSQSAIHLANH 363
Query: 950 PILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWADIFTK 992
+ H R KHI+++ H+IRD ++ + +E + ++ AD+FTK
Sbjct: 364 QVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 109 bits (272), Expect = 6e-24
Identities = 76/246 (30%), Positives = 126/246 (50%), Gaps = 3/246 (1%)
Frame = -3
Query: 84 NGSVLFSGKRKNYIYKIKSSELLSQKVKCLMSVN---DEQWIWLRRLGHASLRKISQLSK 140
+ ++ G K +Y ++ + +S KC + + ++ +W RLGH R
Sbjct: 704 SSQLIGEGVTKGDLYMLEKLDPVSN-YKCSFTSSSSLNKDALWHARLGHPHGRA------ 546
Query: 141 LNLIRGLPRLKYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIG 200
LNL+ LP + + ++ CEAC GK K F + V +L++ DL+ + S
Sbjct: 545 LNLM--LPGVVFENKN-CEACILGKHCKNVFPRTSTVYENC-FDLIYTDLW-TAPSLSRD 381
Query: 201 GKKYGLVIVDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENK 260
KY + +D+ S++TW+ + KD F NF V + + +RSD+GGE+ +
Sbjct: 380 NHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYTSY 201
Query: 261 AFEELFNSQGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAY 320
AF+ + GI H SCP TPQQNGV +RKN+ L E+AR++M +++ ++TA
Sbjct: 200 AFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKHLMEVARSLMFQAN---------VSTAC 48
Query: 321 YIHNII 326
Y+ N I
Sbjct: 47 YLINWI 30
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 105 bits (262), Expect = 9e-23
Identities = 54/132 (40%), Positives = 75/132 (55%)
Frame = +1
Query: 261 AFEELFNSQGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAY 320
+F N + + N TPQQNGV ER NRTL E R M++ + MAK WAEA+ TA
Sbjct: 28 SF*HFVNKRVL*GNSXVAHTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTAC 207
Query: 321 YIHNIISIRPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLG 380
Y+ N I KTP E+ KG+ D S H FG Y++ ++ K D K+ KC FLG
Sbjct: 208 YVINRSPSTVIDLKTPMEMWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLG 387
Query: 381 YSERSKGFRIYN 392
Y++ KG+ +++
Sbjct: 388 YADNVKGYXLWD 423
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 90.1 bits (222), Expect = 4e-18
Identities = 41/87 (47%), Positives = 61/87 (69%)
Frame = +2
Query: 537 LMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFSPVARLEAI 596
L+ KP G IG +W+++ K NE G + + KARLVA+GY +Q+GID+ E F+PV R+E I
Sbjct: 65 LVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPVVRIETI 244
Query: 597 RLLISFSVNHNITLHQMDVKSAFLNGY 623
LL++ + + +H +DVK AFLNG+
Sbjct: 245 *LLLALAATNGC*IHHIDVKIAFLNGH 325
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 89.7 bits (221), Expect = 5e-18
Identities = 45/122 (36%), Positives = 73/122 (58%), Gaps = 3/122 (2%)
Frame = +1
Query: 808 RPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMY---RKTSEYTLSGFCDA 864
RPDI +SV + ++F DPR+ HL A RIL+Y++GT GL++ K+ Y L + D+
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 865 DFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHL 924
D+ GD R+STSG ++W +K+Q ALS+ EAEY++ Q +W+ + +
Sbjct: 301 DWCGD---RRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 925 ED 926
++
Sbjct: 472 KE 477
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 71.2 bits (173), Expect = 2e-12
Identities = 42/125 (33%), Positives = 67/125 (53%), Gaps = 3/125 (2%)
Frame = +1
Query: 657 QAPRAWYERLSSFLLQNEFVRGKGDNTLFC---ITYKNDILIVQIYVDDIIFGSANPSLC 713
Q+PR W++R + + + +++ + D+ +F T K ILIV YVDDI +
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIV--YVDDIFLTGDHGK*I 174
Query: 714 KEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYTLELLKKFNMSDCNISKTP 773
K L+ EFE+ +G LKYFLG+++ + + I Q+KY L+LLK+ M C + P
Sbjct: 175 KRLKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Query: 774 MHPTC 778
C
Sbjct: 355 YGCNC 369
Score = 36.2 bits (82), Expect = 0.066
Identities = 20/56 (35%), Positives = 32/56 (56%)
Frame = +2
Query: 766 DCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARF 821
D S+TPM T L + + V + Y+ ++G L+YL+ +RPDI F V ++F
Sbjct: 332 DVKPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQF 499
>BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T15F17.l
{Arabidopsis thaliana}, partial (3%)
Length = 539
Score = 68.6 bits (166), Expect = 1e-11
Identities = 36/104 (34%), Positives = 58/104 (55%)
Frame = -3
Query: 802 LYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGF 861
+ LT P+I FS++L +R+ S P H +K I KYLKG ++GL Y K L G+
Sbjct: 531 ILLTLQGPNITFSINLLSRYSSAPTMRH*NGIKHICKYLKGIIDMGLFYSKDCSPDLIGY 352
Query: 862 CDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEA 905
+A + D + +S +G G+ +++W S + +TIA S+ A
Sbjct: 351 VNA*YLSDPHKARS*TGYIFTCGNTVISWRSTK*STIATSSNHA 220
>BE941052 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 480
Score = 64.3 bits (155), Expect = 2e-10
Identities = 29/61 (47%), Positives = 41/61 (66%)
Frame = +2
Query: 938 CDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWADIFTKPLAED 997
CD +A L+ NP+ HSR KHI + H++RD VQ+G L +++V T Q AD TKPL++
Sbjct: 29 CDYLSATYLTHNPVYHSRMKHISIDIHFVRDLVQQGKLKVQHVCTVDQLADCLTKPLSKS 208
Query: 998 R 998
R
Sbjct: 209 R 211
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 63.9 bits (154), Expect = 3e-10
Identities = 34/74 (45%), Positives = 46/74 (61%)
Frame = -2
Query: 587 FSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPDHVY 646
F P+ +L I L+S N+ L +DVK+AFL G + E++Y+ QP GF + V
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*E-VGKMVG 378
Query: 647 KLKKSLYGLKQAPR 660
KLKKS+YGLKQ PR
Sbjct: 377 KLKKSMYGLKQGPR 336
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 63.9 bits (154), Expect = 3e-10
Identities = 34/109 (31%), Positives = 58/109 (53%)
Frame = -2
Query: 319 AYYIHNIISIRPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYF 378
A Y+ N I R + ++ P+E+ R+P ++Y FG CY+L E K ++++ K F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 379 LGYSERSKGFRIYNIIHQTVEESIQIRFDGKLGSEKSKLFERFADLSID 427
+GYS KG++ Y+ + V S ++F + G + K E DL+ D
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIEERGYYEEKNQEDLRDLTSD 378
>BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440.1
[imported] - Arabidopsis thaliana, partial (20%)
Length = 731
Score = 58.2 bits (139), Expect = 2e-08
Identities = 29/84 (34%), Positives = 49/84 (57%), Gaps = 1/84 (1%)
Frame = +3
Query: 912 TCCTQTIWMKNHLEDYGLS-LKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHV 970
T Q +W+++ L + ++V I DN + I+L++NP+ H R HI +YH+IR+ V
Sbjct: 123 TAARQAMWLQDLLSEVTWEPCEEVVIRIDNQSVIALTRNPVFHGRGNHIHKRYHFIRECV 302
Query: 971 QKGTLSLEYVDTDHQWADIFTKPL 994
+ G + +E+V + A I TK L
Sbjct: 303 ENGQVEVEHVPGEKHRAYI*TKAL 374
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,884,338
Number of Sequences: 36976
Number of extensions: 456825
Number of successful extensions: 2254
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 2217
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2241
length of query: 1017
length of database: 9,014,727
effective HSP length: 106
effective length of query: 911
effective length of database: 5,095,271
effective search space: 4641791881
effective search space used: 4641791881
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0166.5


