

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0166.12
(1518 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
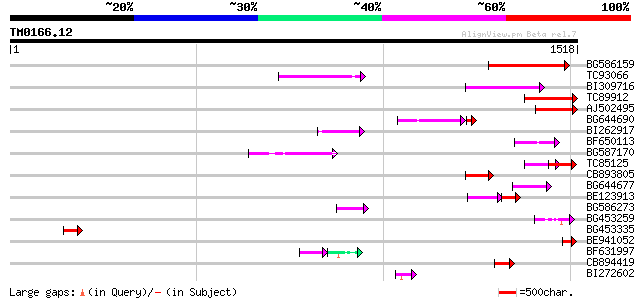
Score E
Sequences producing significant alignments: (bits) Value
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 188 1e-47
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 132 1e-30
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 116 8e-26
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 115 1e-25
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 115 1e-25
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 104 1e-24
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 96 8e-20
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 96 8e-20
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 89 1e-17
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 73 1e-12
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 72 2e-12
BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T... 70 5e-12
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 47 1e-10
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 61 3e-09
BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, ... 48 3e-05
BG453335 weakly similar to GP|18071369|g putative gag-pol polypr... 47 4e-05
BE941052 weakly similar to PIR|B85188|B85 retrotransposon like p... 45 2e-04
BF631997 weakly similar to GP|18542925|gb Putative pol polyprote... 38 2e-04
CB894419 weakly similar to GP|10177935|db copia-type polyprotein... 45 2e-04
BI272602 GP|11602812|db avenaIII {Gallus gallus}, partial (3%) 44 4e-04
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 188 bits (478), Expect = 1e-47
Identities = 91/217 (41%), Positives = 139/217 (63%), Gaps = 1/217 (0%)
Frame = +1
Query: 1283 LQVKQMQDSIFISQSKYAK*LVKKFGLEGSTHKRTPAATHIKLTKDESGTDVDQSLYRSM 1342
++V Q ++ I+I Q KY L+++FG+E S R P A KL KDE+G VD + Y+ +
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 1343 ICSLLYLTASRPDIAFSIDVCARYQAAPKESHLIHVKRIIKYINGTTDYGTFYAHNSNSN 1402
+ L+YL A+RPD+ + + + +R+ P E H+ VKR+++Y+NGT + G Y N +
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 1403 LISYCDADWAGNADDGKSTSGGCFFLGNNLISWFNKKQNCVSLSTTEAEYIAAGSWCTQL 1462
L +Y D+D+AG+ DD KSTSG F L + +SW +KKQ V+LSTT+AE+IAA Q
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 1463 LWMKHMLREYNVKQD-VMTLYCENLSAINISKNPIQH 1498
+WM+ +L + Q +T+YC+N S I +SKNP+ H
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 132 bits (332), Expect = 1e-30
Identities = 83/236 (35%), Positives = 119/236 (50%), Gaps = 2/236 (0%)
Frame = +1
Query: 720 GKQTKTSHKKLTQIGTTRSLQLLHMDLMGPMHKESFGGKRYVFVCVDDFSRFTWVNFLAE 779
G + K S T T L +H DL GP S+GG+RY+ +DDF R WV FL
Sbjct: 43 GNRKKVSFSTATH-RTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRY 219
Query: 780 KSETFEVFRELSQQLQGEKGYGIVKIRSDHGKEFENGKFSEYCGNEGIKHEFSAPITPQQ 839
K+ETF F++ ++ + G + K+ +D+ EF + F+E+C N GI + P PQQ
Sbjct: 220 KNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQ 399
Query: 840 NGVVERKNRTLQESARVMLHAKKV--PYHLWAEAINTACYIHYRVTIRSGTDSTLYELWK 897
NGV ER RTL E AR ML + LW EA +TAC++ R + ++W
Sbjct: 400 NGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWS 579
Query: 898 GKKPTVKYFYVFESRCYILTDREQRRKLDPKSDEGLFLGYSINSKAYRVFNSRTKT 953
G +F Y L + KL P++ E +FL Y+ SK YR++ S K+
Sbjct: 580 GNLVDYSNLRIFGCPAYALVN---DGKLAPRAGECIFLSYASESKGYRLWCSDPKS 738
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 116 bits (290), Expect = 8e-26
Identities = 66/211 (31%), Positives = 107/211 (50%)
Frame = +2
Query: 1220 YNKRGIDKTLFVKSDGGKVMIAQIYVDDIVFGGMSDHMVEHFVRHMKSEFEMSLVGELNY 1279
Y + D +LF K +YVDDIV G ++H + F++ +G L Y
Sbjct: 110 YLQSSSDFSLFTKFKDSSFTTLLVYVDDIVLAGNDISEIQHVKCFLIDRFKIKDLGSLRY 289
Query: 1280 FPGLQVKQMQDSIFISQSKYAK*LVKKFGLEGSTHKRTPAATHIKLTKDESGTDVDQSLY 1339
F GL+V + + I ++Q KY L++ G TP +KL +S D++ Y
Sbjct: 290 FLGLEVARSKQGILLNQRKYTLELLEDSGNLAVKSTLTPYDISLKLHNSDSPLYNDETQY 469
Query: 1340 RSMICSLLYLTASRPDIAFSIDVCARYQAAPKESHLIHVKRIIKYINGTTDYGTFYAHNS 1399
R +I L+YLT +RPDI+F++ +++ + P++ H R+++Y+ G FY+ S
Sbjct: 470 RRLIGKLIYLTTTRPDISFAVQQLSQFVSKPQQVHYQAAIRVLQYLKTAPAKGLFYSATS 649
Query: 1400 NSNLISYCDADWAGNADDGKSTSGGCFFLGN 1430
N L S+ D+DWA KS +G FLG+
Sbjct: 650 NLKLSSFADSDWATCPTTRKSVTGYWVFLGS 742
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 115 bits (289), Expect = 1e-25
Identities = 59/143 (41%), Positives = 87/143 (60%), Gaps = 2/143 (1%)
Frame = +1
Query: 1378 VKRIIKYINGTTDYGTFY--AHNSNSNLISYCDADWAGNADDGKSTSGGCFFLGNNLISW 1435
+K ++KY+N + Y A L Y DAD+AGN D KS SG F L ISW
Sbjct: 7 LKWVLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTISW 186
Query: 1436 FNKKQNCVSLSTTEAEYIAAGSWCTQLLWMKHMLREYNVKQDVMTLYCENLSAINISKNP 1495
+Q+ V+LSTT+AEYIA +W+K M+ E + Q+ + ++C++ SAI+++ +
Sbjct: 187 KANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGITQEYVKIHCDSQSAIHLANHQ 366
Query: 1496 IQHSRTKHIDIRHHFIKDLVEEK 1518
+ H RTKHIDIR HFI+D++E K
Sbjct: 367 VYHERTKHIDIRLHFIRDMIESK 435
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 115 bits (288), Expect = 1e-25
Identities = 56/111 (50%), Positives = 78/111 (69%), Gaps = 1/111 (0%)
Frame = +2
Query: 1409 ADWAGNADDGKSTSGGCFFLGNNLISWFNKKQNCVSLSTTEAEYIAAGSWCTQLLWMKHM 1468
+DWAG+ + KSTSG F LG ISW +KKQ V+ ST EAEYIA+ S TQ +W++ +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 1469 LREYNVKQDVMT-LYCENLSAINISKNPIQHSRTKHIDIRHHFIKDLVEEK 1518
L + +Q+ T +YC+N SAI +SKNP+ H R+KHIDI+ H I++L+ EK
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEK 334
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 104 bits (260), Expect(2) = 1e-24
Identities = 73/181 (40%), Positives = 109/181 (59%)
Frame = -3
Query: 1039 RDIIANACFISKIEPKNIKEALTDEYWINAMQEELSQFKRNEVWDLVPRPEVVM*LVPSG 1098
R++++ + FIS IEPKN+KEAL D WIN+MQEEL QF+R++VW LVPRPE ++ +
Sbjct: 627 RNLVSFSAFISSIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKT-VIGTR 451
Query: 1099 FSETSQMKVIL*QETRPD*LLKGILRLKELILMRPLLMLLDWNQSDCC*D*HAF*NSNCS 1158
+ ++++ +L E P K ++ KE +MR +L +W + * S+C+
Sbjct: 450 WVFRNKLE*LL--EINPSWWCKDTIKKKE*TMMRLFHLLPEWKLLEF**LLLHSWGSSCT 277
Query: 1159 RWM*RVLS*MDT*MRKSM*NNQRAL*IQPFQNMFTS*ERRYMD*SKSQGLDMEG*QSF** 1218
+WM*RV M+ R+ + +N L +Q +Q M + * R YM *SK Q M+G QSF *
Sbjct: 276 KWM*RVHLLMEISKRRCLSSNLLDLKMQRYQIMCSD*IRHYMV*SKLQEHGMKGCQSFC* 97
Query: 1219 R 1219
R
Sbjct: 96 R 94
Score = 28.5 bits (62), Expect(2) = 1e-24
Identities = 15/28 (53%), Positives = 20/28 (70%), Gaps = 1/28 (3%)
Frame = -2
Query: 1222 KRG-IDKTLFVKSDGGKVMIAQIYVDDI 1248
KRG ID TLF+ +++I Q+YVDDI
Sbjct: 85 KRGKIDNTLFLLKRE*ELLIIQVYVDDI 2
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 96.3 bits (238), Expect = 8e-20
Identities = 52/126 (41%), Positives = 69/126 (54%)
Frame = +1
Query: 823 GNEGIKHEFSAPITPQQNGVVERKNRTLQESARVMLHAKKVPYHLWAEAINTACYIHYRV 882
GN + H TPQQNGV ER NRTL E R ML + WAEA+ TACY+ R
Sbjct: 64 GNSXVAH------TPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRS 225
Query: 883 TIRSGTDSTLYELWKGKKPTVKYFYVFESRCYILTDREQRRKLDPKSDEGLFLGYSINSK 942
T E+WKGK +VF Y++ + ++R KLDPKS + +FLGY+ N K
Sbjct: 226 PSTVIDLKTPMEMWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVK 405
Query: 943 AYRVFN 948
Y +++
Sbjct: 406 GYXLWD 423
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 96.3 bits (238), Expect = 8e-20
Identities = 49/122 (40%), Positives = 72/122 (58%), Gaps = 3/122 (2%)
Frame = +1
Query: 1353 RPDIAFSIDVCARYQAAPKESHLIHVKRIIKYINGTTDYGTFYAHNSNS---NLISYCDA 1409
RPDI +S+ V +++ P++ HLI RI++Y+ GT +YG + + + S LI Y D+
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 1410 DWAGNADDGKSTSGGCFFLGNNLISWFNKKQNCVSLSTTEAEYIAAGSWCTQLLWMKHML 1469
DW G D +STSG F + ISW KKQ +LS+ EAEYIA Q LW+ ++
Sbjct: 301 DWCG---DRRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 1470 RE 1471
+E
Sbjct: 472 KE 477
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 89.0 bits (219), Expect = 1e-17
Identities = 64/243 (26%), Positives = 111/243 (45%), Gaps = 3/243 (1%)
Frame = -3
Query: 639 ENEEVVMTGVRSKDNCYMWS---PQNKAMSSMCLVSMEDEAKLWHQK*GHLNPTSMKKVI 695
E+ +++ GV +K + YM P + S S ++ LWH + GH + ++ ++
Sbjct: 707 ESSQLIGEGV-TKGDLYMLEKLDPVSNYKCSFTSSSSLNKDALWHARLGHPHGRALNLML 531
Query: 696 SKEAIRGIPRLKIEEDKVCGECQIGKQTKTSHKKLTQIGTTRSLQLLHMDLMGPMHKESF 755
G+ + E+K C C +GK K + + + L++ DL S
Sbjct: 530 P-----GV----VFENKNCEACILGKHCKNVFPRTSTV-YENCFDLIYTDLW-TAPSLSR 384
Query: 756 GGKRYVFVCVDDFSRFTWVNFLAEKSETFEVFRELSQQLQGEKGYGIVKIRSDHGKEFEN 815
+Y +D+ S++TW+ + K + F+ + I +RSD+G E+ +
Sbjct: 383 DNHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYTS 204
Query: 816 GKFSEYCGNEGIKHEFSAPITPQQNGVVERKNRTLQESARVMLHAKKVPYHLWAEAINTA 875
F + + GI H+ S P TPQQNGV +RKN+ L E AR ++ V +TA
Sbjct: 203 YAFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKHLMEVARSLMFQANV---------STA 51
Query: 876 CYI 878
CY+
Sbjct: 50 CYL 42
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 72.8 bits (177), Expect = 1e-12
Identities = 29/76 (38%), Positives = 53/76 (69%)
Frame = +3
Query: 1442 CVSLSTTEAEYIAAGSWCTQLLWMKHMLREYNVKQDVMTLYCENLSAINISKNPIQHSRT 1501
C S++ + + C + +WM+ ++ E KQ+ +T+YC++ SA++I++NP HSRT
Sbjct: 192 CSSVNDRSGVHGSLPQACKEAIWMQRLMEELGHKQEQITVYCDSQSALHIARNPAFHSRT 371
Query: 1502 KHIDIRHHFIKDLVEE 1517
KHI I++HF++++VEE
Sbjct: 372 KHIGIQYHFVREVVEE 419
Score = 68.9 bits (167), Expect = 1e-11
Identities = 39/98 (39%), Positives = 56/98 (56%)
Frame = +1
Query: 1378 VKRIIKYINGTTDYGTFYAHNSNSNLISYCDADWAGNADDGKSTSGGCFFLGNNLISWFN 1437
VKRI++YI GT+ + S + Y D+D+AG+ D KST+G F L +SW +
Sbjct: 1 VKRIMRYIKGTSGVAVCFG-GSELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLS 177
Query: 1438 KKQNCVSLSTTEAEYIAAGSWCTQLLWMKHMLREYNVK 1475
K Q V+LSTTEAEY+AA ++KH + + K
Sbjct: 178 KLQTVVALSTTEAEYMAA--------YLKHARKLFGCK 267
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 72.0 bits (175), Expect = 2e-12
Identities = 36/81 (44%), Positives = 55/81 (67%), Gaps = 6/81 (7%)
Frame = +3
Query: 1220 YNKRGIDK-----TLFVK-SDGGKVMIAQIYVDDIVFGGMSDHMVEHFVRHMKSEFEMSL 1273
+ K G +K TLFVK S+GGK++I +YVDD++F G ++M E F + MK EF MS
Sbjct: 531 FTKEGFEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSD 710
Query: 1274 VGELNYFPGLQVKQMQDSIFI 1294
+G+++YF G++V Q + I+I
Sbjct: 711 LGKMHYFLGVEVTQNEKGIYI 773
Score = 30.0 bits (66), Expect = 7.1
Identities = 11/33 (33%), Positives = 18/33 (54%)
Frame = +3
Query: 1052 EPKNIKEALTDEYWINAMQEELSQFKRNEVWDL 1084
+P +EA+ E W +M E+ +RN W+L
Sbjct: 39 DPTTFEEAVKSEKWRASMNNEMEATERNNTWEL 137
>BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T15F17.l
{Arabidopsis thaliana}, partial (3%)
Length = 539
Score = 70.5 bits (171), Expect = 5e-12
Identities = 36/104 (34%), Positives = 59/104 (56%)
Frame = -3
Query: 1347 LYLTASRPDIAFSIDVCARYQAAPKESHLIHVKRIIKYINGTTDYGTFYAHNSNSNLISY 1406
+ LT P+I FSI++ +RY +AP H +K I KY+ G D G FY+ + + +LI Y
Sbjct: 531 ILLTLQGPNITFSINLLSRYSSAPTMRH*NGIKHICKYLKGIIDMGLFYSKDCSPDLIGY 352
Query: 1407 CDADWAGNADDGKSTSGGCFFLGNNLISWFNKKQNCVSLSTTEA 1450
+A + + +S +G F GN +ISW + K + ++ S+ A
Sbjct: 351 VNA*YLSDPHKARS*TGYIFTCGNTVISWRSTK*STIATSSNHA 220
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 47.0 bits (110), Expect(2) = 1e-10
Identities = 31/94 (32%), Positives = 46/94 (47%), Gaps = 1/94 (1%)
Frame = +1
Query: 1226 DKTLFVK-SDGGKVMIAQIYVDDIVFGGMSDHMVEHFVRHMKSEFEMSLVGELNYFPGLQ 1284
D +F+K S K I +YVDDI G ++ + EFE+ +G L YF G++
Sbjct: 73 DHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKRLKNLLAEEFEIKDLGNLKYFLGME 252
Query: 1285 VKQMQDSIFISQSKYAK*LVKKFGLEGSTHKRTP 1318
V + + ISQ KY L+K+ + G R P
Sbjct: 253 VARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Score = 38.5 bits (88), Expect(2) = 1e-10
Identities = 17/50 (34%), Positives = 32/50 (64%)
Frame = +2
Query: 1317 TPAATHIKLTKDESGTDVDQSLYRSMICSLLYLTASRPDIAFSIDVCARY 1366
TP +KL ++GT VD+ Y+ ++ L+YL+ +RPDI+F + +++
Sbjct: 350 TPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQF 499
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 61.2 bits (147), Expect = 3e-09
Identities = 28/86 (32%), Positives = 48/86 (55%)
Frame = -2
Query: 875 ACYIHYRVTIRSGTDSTLYELWKGKKPTVKYFYVFESRCYILTDREQRRKLDPKSDEGLF 934
ACY+ R+ R D +E+ +KP++ Y VF CY+L E R KL+ +S + +F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 935 LGYSINSKAYRVFNSRTKTMMESINV 960
+GYS K Y+ ++ + ++ S +V
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDV 447
>BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, partial (5%)
Length = 657
Score = 47.8 bits (112), Expect = 3e-05
Identities = 41/114 (35%), Positives = 54/114 (46%), Gaps = 8/114 (7%)
Frame = -2
Query: 1406 YCDADWAGNADDGKSTSGGCFFLGNNLISWFNKKQNCVSLSTTEAEYIAAGSWCTQLLWM 1465
Y + AG D STSG FLG N++ KQN V+ + C+Q L
Sbjct: 491 YXNTVCAGWIVDRGSTSGY*MFLGGNMVE*---KQNVVAR*VQRHNF----ELCSQGL*- 336
Query: 1466 KHMLREYNVK--------QDVMTLYCENLSAINISKNPIQHSRTKHIDIRHHFI 1511
+ M E +K +D MTL+ N I+ NP+QH RTKHI+I HFI
Sbjct: 335 RVMDEELKIKLDDLIINYKDPMTLF*NNNFVSRIAHNPVQHYRTKHIEIDQHFI 174
>BG453335 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (11%)
Length = 660
Score = 47.4 bits (111), Expect = 4e-05
Identities = 21/51 (41%), Positives = 37/51 (72%)
Frame = +1
Query: 145 QKILRSLPKKFDMKVTAIEEAQDIGSIKVDELIGSLQTFEMSVNERSDKKS 195
+KI+R+L +FD V AI E++D+ ++K++EL SL+ ++ V ERS ++S
Sbjct: 409 EKIMRTLSPRFDFIVVAIHESKDVKTLKIEELKSSLEAHKLMVYERSSERS 561
>BE941052 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 480
Score = 45.4 bits (106), Expect = 2e-04
Identities = 19/37 (51%), Positives = 27/37 (72%)
Frame = +2
Query: 1481 LYCENLSAINISKNPIQHSRTKHIDIRHHFIKDLVEE 1517
L C+ LSA ++ NP+ HSR KHI I HF++DLV++
Sbjct: 23 LRCDYLSATYLTHNPVYHSRMKHISIDIHFVRDLVQQ 133
>BF631997 weakly similar to GP|18542925|gb Putative pol polyprotein {Oryza
sativa}, partial (6%)
Length = 650
Score = 38.1 bits (87), Expect(2) = 2e-04
Identities = 23/73 (31%), Positives = 37/73 (50%)
Frame = -2
Query: 777 LAEKSETFEVFRELSQQLQGEKGYGIVKIRSDHGKEFENGKFSEYCGNEGIKHEFSAPIT 836
+ KSE F R + ++G + ++G E + +F+E C E I +++ T
Sbjct: 637 MKHKSEAFYFSRNGRFWSRVKQGRR*NVFKLNNGLEICSAEFNELCKEEHITRQYTVRNT 458
Query: 837 PQQNGVVERKNRT 849
PQ+NGV ER N T
Sbjct: 457 PQKNGVAERMNIT 419
Score = 26.2 bits (56), Expect(2) = 2e-04
Identities = 29/101 (28%), Positives = 41/101 (39%), Gaps = 9/101 (8%)
Frame = -1
Query: 852 ESARVMLHAKKVPYHLWAEAINTACYI---------HYRVTIRSGTDSTLYELWKGKKPT 902
E AR M + AEAI+T CY+ R+ +R G + L G
Sbjct: 413 ERARCMFSNAGLNRSFQAEAISTKCYLVNVLPLLL*TVRLHLRYGLINLLITQILG---C 243
Query: 903 VKYFYVFESRCYILTDREQRRKLDPKSDEGLFLGYSINSKA 943
Y++V E KL+P+S +GLF G + KA
Sbjct: 242 PAYYHVNEG------------KLEPRSKKGLFWGKEMV*KA 156
>CB894419 weakly similar to GP|10177935|db copia-type polyprotein {Arabidopsis
thaliana}, partial (2%)
Length = 170
Score = 45.1 bits (105), Expect = 2e-04
Identities = 24/55 (43%), Positives = 34/55 (61%)
Frame = +3
Query: 1298 KYAK*LVKKFGLEGSTHKRTPAATHIKLTKDESGTDVDQSLYRSMICSLLYLTAS 1352
K A ++KKF +E S TP +KLT++ G VD + Y+S+I SL YLTA+
Sbjct: 6 KCASDILKKFKMEHSKPISTPVEEKLKLTRESDGKRVDSTHYKSLIGSLRYLTAT 170
>BI272602 GP|11602812|db avenaIII {Gallus gallus}, partial (3%)
Length = 488
Score = 44.3 bits (103), Expect = 4e-04
Identities = 23/63 (36%), Positives = 32/63 (50%), Gaps = 8/63 (12%)
Frame = +1
Query: 1033 GVVTRSRDIIANACF--------ISKIEPKNIKEALTDEYWINAMQEELSQFKRNEVWDL 1084
G++TRS++ I + IEP NI +AL D W + MQ+E + N WDL
Sbjct: 187 GIITRSQNNIVKTIKKLNLHVRPLCPIEPSNITQALRDHDWRSIMQDEFDALQNNNTWDL 366
Query: 1085 VPR 1087
V R
Sbjct: 367 VSR 375
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.329 0.141 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,642,371
Number of Sequences: 36976
Number of extensions: 643982
Number of successful extensions: 3941
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 2872
Number of HSP's successfully gapped in prelim test: 148
Number of HSP's that attempted gapping in prelim test: 978
Number of HSP's gapped (non-prelim): 3120
length of query: 1518
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1409
effective length of database: 4,984,343
effective search space: 7022939287
effective search space used: 7022939287
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0166.12


