

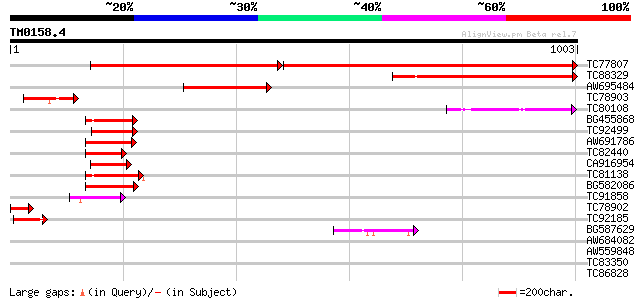
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0158.4
(1003 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77807 similar to PIR|T52601|T52601 squamosa promoter binding p... 910 0.0
TC88329 similar to PIR|T47827|T47827 squamosa promoter binding p... 518 e-147
AW695484 similar to PIR|T52602|T526 squamosa promoter binding pr... 227 2e-59
TC78903 134 2e-31
TC80108 similar to GP|22045251|gb|AAL69483.2 putative SPL1-relat... 133 4e-31
BG455868 similar to SP|Q38741|SBP1_ Squamosa-promoter binding pr... 123 3e-28
TC92499 homologue to PIR|T52594|T52594 squamosa promoter binding... 122 5e-28
AW691786 similar to PIR|T52298|T52 squamosa promoter binding pro... 119 8e-27
TC82440 similar to PIR|H96793|H96793 unknown protein F14G6.18 [i... 107 3e-23
CA916954 similar to PIR|T52592|T525 squamosa-promoter binding pr... 103 4e-22
TC81138 similar to PIR|T52606|T52606 squamosa promoter binding p... 100 2e-21
BG582086 similar to PIR|T52603|T526 squamosa promoter binding pr... 100 3e-21
TC91858 similar to PIR|T52592|T52592 squamosa-promoter binding p... 90 5e-18
TC78902 67 4e-11
TC92185 similar to GP|13374857|emb|CAC34491. putative protein {A... 63 5e-10
BG587629 similar to PIR|T52605|T526 squamosa promoter binding pr... 58 2e-08
AW684082 homologue to PIR|T47773|T47 hypothetical protein F24I3.... 35 0.14
AW559848 homologue to GP|21592490|gb| unknown {Arabidopsis thali... 33 0.71
TC83350 similar to GP|21555074|gb|AAM63770.1 unknown {Arabidopsi... 32 1.6
TC86828 similar to PIR|T08454|T08454 hypothetical protein F22O6.... 30 3.5
>TC77807 similar to PIR|T52601|T52601 squamosa promoter binding protein 1
[imported] - Arabidopsis thaliana, partial (55%)
Length = 2923
Score = 910 bits (2352), Expect(2) = 0.0
Identities = 450/523 (86%), Positives = 481/523 (91%), Gaps = 3/523 (0%)
Frame = +1
Query: 484 SPTDIESYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGWVHIRVQ 543
SPTDIESYIRPGCIVLTIYLRQAEAVWEE+C DLTSSL +LLDVSDDTFW+TGWVHIRVQ
Sbjct: 1024 SPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRVQ 1203
Query: 544 HQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIRPATRLMCA 603
HQMAFIFNGQVVIDTSLPFRSNNYSKI TVSPIA PASKR QFSVKGVN++RPATRLMCA
Sbjct: 1204 HQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPATRLMCA 1383
Query: 604 LEGKYLVCEDAHESMDQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGLSSSFFPFIVA 663
LEGKYLVCEDAHES DQYSEELD L CIQFSC+VPV+NGRGFIEIEDQGLSSSFFPFIVA
Sbjct: 1384 LEGKYLVCEDAHESTDQYSEELDELQCIQFSCSVPVSNGRGFIEIEDQGLSSSFFPFIVA 1563
Query: 664 EEDVCSDICGLEPLLELSETDPDIEGTGKIKAKSQAMDFIHEIGWLLHGSQLKSRMVHLS 723
EEDVC++I LEPLLE SETDPDIEGTGKIKAKSQAMDFIHE+GWLLH SQLK RMV+L+
Sbjct: 1564 EEDVCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQLKYRMVNLN 1743
Query: 724 SSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLEGTVSTGDHPSLDLALSEMGLLHRA 783
S DLFPL+RF WLMEFSMDHDWCAVVKKLLNLLL+ TV+ GDHP+L ALSEMGLLHRA
Sbjct: 1744 SGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGDHPTLYQALSEMGLLHRA 1923
Query: 784 VRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPAGLTPLHIAAGK 843
VRRNSKQLVELLL YVP+N S+E+ PE KALV G+ S+LFRPDA GPAGLTPLHIAAGK
Sbjct: 1924 VRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAGK 2103
Query: 844 DGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKRQGAP 903
DGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINK QGA
Sbjct: 2104 DGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGAA 2283
Query: 904 HVVVEIPTNLTENDTNQKQNESSTTFEIAKT---RDQGHCKLCDIKLSCRTAVGRSLVYK 960
HVVVEIP+N+TE++ N KQNES T+ EI K R QG+CKLCD K+SCRTAVGRS+VY+
Sbjct: 2284 HVVVEIPSNMTESNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVYR 2463
Query: 961 PAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGTS 1003
PAMLSMVA+AAVCVCVALLFKSSPEVLYMFRPF WESLDFGTS
Sbjct: 2464 PAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGTS 2592
Score = 508 bits (1309), Expect(2) = 0.0
Identities = 259/340 (76%), Positives = 283/340 (83%), Gaps = 1/340 (0%)
Frame = +3
Query: 144 SNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFD 203
SNRAVCQVEDC ADLSR KDYHRRHKVCEMHSKA+RALVGNAMQRFCQQCSRFH+L+EFD
Sbjct: 3 SNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFD 182
Query: 204 EGKRSCRRRLAGHNKRRRKTNNEAVPNGNPLNDDQTSSYLLISLLKILSNMNSDRSDQTT 263
EGKRSCRRRLAGHNKRRRKTN EAVPNG+P NDDQTSSYLLISLLKILSNM+SDRSDQ T
Sbjct: 183 EGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLISLLKILSNMHSDRSDQPT 362
Query: 264 DQDLLTHLLRSLASRNGEQGGNNLSNILREPENLLREGGSSRESEMVPTLLSNGSQGSPT 323
DQDLLTHLLRSLAS+N EQG NLSN+LRE ENLLREGGSSR S MV L SNGSQGSPT
Sbjct: 363 DQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPT 542
Query: 324 DIRQHQTVSMNKMQQEVVHAHDARATDQQLMSYIKPSISNTPPAYAEARDSTAGQIKMNN 383
I QHQ VSMN+MQQE+VH HD R +D QL+S IKPSISN+PPAY+E RDS +GQ KMNN
Sbjct: 543 VITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDS-SGQTKMNN 719
Query: 384 FDLNDIYIDSDDGIEDLERLPGTTNHVTSSLDYPWTQQDSHQSSPPQTSRNSESGSAHSP 443
FDLNDIY+DSDDG EDLERLP +TN TSS+DYPWTQQDSHQSSP QTS NS+S SA P
Sbjct: 720 FDLNDIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQVP 899
Query: 444 S-SSTGEAQCRTDRIVIKLFGKEPSDFPLILRAQILDWLS 482
+ + + + GK +F L L +ILDWLS
Sbjct: 900 F*F*RKKPRVAQTELFLNSSGKSQMNFLLYLEHRILDWLS 1019
>TC88329 similar to PIR|T47827|T47827 squamosa promoter binding protein-like
12 [imported] - Arabidopsis thaliana, partial (19%)
Length = 1277
Score = 518 bits (1334), Expect = e-147
Identities = 259/330 (78%), Positives = 290/330 (87%), Gaps = 3/330 (0%)
Frame = +3
Query: 677 LLELSETDPDIEGTGKIKAKSQAMDFIHEIGWLLHGSQLKSRMVHLSSSTDLFPLKRFKW 736
LLE S+T PD EG GKI+AK+QAMDFIHE+GWLLH Q+KS V L+SS DLFPL RFKW
Sbjct: 3 LLESSDTYPDNEGAGKIQAKNQAMDFIHEMGWLLHRRQIKSS-VRLNSSMDLFPLDRFKW 179
Query: 737 LMEFSMDHDWCAVVKKLLNLLLEGTVSTGDHPSLDLALSEMGLLHRAVRRNSKQLVELLL 796
LMEFS+DHDWCAVVKKLLNL+L+GTVSTGDH SL LALSE+GLLHRAVRRNS+QLVELLL
Sbjct: 180 LMEFSVDHDWCAVVKKLLNLMLDGTVSTGDHTSLYLALSELGLLHRAVRRNSRQLVELLL 359
Query: 797 TYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPAGLTPLHIAAGKDGSEDVLDALTND 856
+VP+NISN++ PE KALV GE Q+FLFRPDA GPAGLTPLHIAAGKDGSEDVLDALTND
Sbjct: 360 RFVPQNISNKLGPEDKALVNGENQNFLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTND 539
Query: 857 PCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKRQGAPHVVVEIPTNLTEN 916
PCMVGIEAW +ARDSTGSTPEDYARLRGHYTYIHLVQKKINK QG HVVV+IP+ T+
Sbjct: 540 PCMVGIEAWNSARDSTGSTPEDYARLRGHYTYIHLVQKKINKSQGGAHVVVDIPSIPTKF 719
Query: 917 DTNQKQNESSTTFEIAKT---RDQGHCKLCDIKLSCRTAVGRSLVYKPAMLSMVAVAAVC 973
DT+QK++ES TTF+I + + CKLCD KLSCRTAV +S VY+PAMLSMVA+AAVC
Sbjct: 720 DTSQKKDESCTTFQIGNAEVKKVRKDCKLCDHKLSCRTAVRKSFVYRPAMLSMVAIAAVC 899
Query: 974 VCVALLFKSSPEVLYMFRPFSWESLDFGTS 1003
VCVALLFKSSPEVLY+FRPF WESLD+GTS
Sbjct: 900 VCVALLFKSSPEVLYIFRPFRWESLDYGTS 989
>AW695484 similar to PIR|T52602|T526 squamosa promoter binding protein 1
[imported] - Arabidopsis thaliana, partial (8%)
Length = 468
Score = 227 bits (578), Expect = 2e-59
Identities = 113/155 (72%), Positives = 129/155 (82%)
Frame = +2
Query: 308 EMVPTLLSNGSQGSPTDIRQHQTVSMNKMQQEVVHAHDARATDQQLMSYIKPSISNTPPA 367
EMV TL++NGSQGSPT Q+QTVS++++Q +V+H+HDAR DQQ KP +SN+PPA
Sbjct: 2 EMVSTLVTNGSQGSPTVTVQNQTVSISEIQHQVMHSHDARVADQQTTFSAKPGVSNSPPA 181
Query: 368 YAEARDSTAGQIKMNNFDLNDIYIDSDDGIEDLERLPGTTNHVTSSLDYPWTQQDSHQSS 427
Y+EAR STAGQ KMN+FDLNDIYIDSDDGIED+ERLP TTN SSLDYPW QQDSHQSS
Sbjct: 182 YSEARYSTAGQTKMNDFDLNDIYIDSDDGIEDIERLPVTTNLGASSLDYPWMQQDSHQSS 361
Query: 428 PPQTSRNSESGSAHSPSSSTGEAQCRTDRIVIKLF 462
PPQTS NS+S SA SPSSSTGE Q RTDRIV KLF
Sbjct: 362 PPQTSGNSDSASAQSPSSSTGETQNRTDRIVFKLF 466
>TC78903
Length = 332
Score = 134 bits (336), Expect = 2e-31
Identities = 70/103 (67%), Positives = 81/103 (77%), Gaps = 6/103 (5%)
Frame = +3
Query: 25 GKRSAEWDLNNWRWDGDLFIASRVNPVQEDG--VGQQLFPLVSGIP----ASNSSSSCSE 78
GKRS EW+LN+WRWDGDLFIASRVN VQ + VGQQ FPL SGIP +SN+SSSCSE
Sbjct: 3 GKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPLGSGIPVVGGSSNTSSSCSE 182
Query: 79 EVDLRDPMGSREGERKRRVIVLEDDGLNEEGGTLSLKLGGHAA 121
E DL G++EGE+KRRVIVLEDDGLN++ G LSL L GH +
Sbjct: 183 EGDLE--KGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVS 305
>TC80108 similar to GP|22045251|gb|AAL69483.2 putative SPL1-related protein
{Arabidopsis thaliana}, partial (42%)
Length = 1251
Score = 133 bits (334), Expect = 4e-31
Identities = 80/230 (34%), Positives = 130/230 (55%), Gaps = 1/230 (0%)
Frame = +2
Query: 774 LSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPAG 833
L + LL+RAV+R +V+LL+ Y +I+++ + ++F P+ GP G
Sbjct: 29 LKAIQLLNRAVKRKCTSMVDLLINY---SITSK---------NDTSKKYVFPPNLEGPGG 172
Query: 834 LTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQ 893
+TPLH+AA SE V+D+LTNDP +G++ W+ D G TP YA +R +++Y LV
Sbjct: 173 ITPLHLAASTTDSEGVIDSLTNDPQEIGLKCWETLADENGQTPHAYAMMRNNHSYNMLVA 352
Query: 894 KKINKRQGAPHVVVEIPTNLTENDTNQKQNESSTTFEIAKTRDQ-GHCKLCDIKLSCRTA 952
+K + RQ + V V I N E+ + + ++ + D C + +++ R +
Sbjct: 353 RKCSDRQRS-EVSVRI-DNEIEHPSLGIELMQKRINQVKRVGDSCSKCAIAEVRAKRRFS 526
Query: 953 VGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGT 1002
RS ++ P + SM+AVAAVCVCV +LF+ +P V PF WE+L++GT
Sbjct: 527 GSRSWLHGPFIHSMLAVAAVCVCVCVLFRGTPYV-GSVSPFRWENLNYGT 673
>BG455868 similar to SP|Q38741|SBP1_ Squamosa-promoter binding protein 1.
[Garden snapdragon] {Antirrhinum majus}, partial (70%)
Length = 656
Score = 123 bits (309), Expect = 3e-28
Identities = 59/93 (63%), Positives = 69/93 (73%)
Frame = +1
Query: 134 KKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQC 193
KK AGG + CQV++C ADLS AK YH+RHKVCE HSKA L+ QRFCQQC
Sbjct: 226 KKGSKAGGSVTPS--CQVDNCNADLSAAKQYHKRHKVCENHSKAHSVLISELQQRFCQQC 399
Query: 194 SRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNE 226
SRFH + EFD+ KRSCRRRLAGHN+RRRK+ +E
Sbjct: 400 SRFHEVSEFDDLKRSCRRRLAGHNERRRKSASE 498
>TC92499 homologue to PIR|T52594|T52594 squamosa promoter binding protein 8
[imported] - Arabidopsis thaliana, partial (22%)
Length = 805
Score = 122 bits (307), Expect = 5e-28
Identities = 55/81 (67%), Positives = 61/81 (74%)
Frame = +1
Query: 145 NRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDE 204
N CQ E C ADLS+AK YHRRHKVCE HSKA + QRFCQQCSRFH+L EFD
Sbjct: 10 NSPRCQAEGCNADLSQAKHYHRRHKVCEFHSKAATVVAAGLTQRFCQQCSRFHLLSEFDN 189
Query: 205 GKRSCRRRLAGHNKRRRKTNN 225
GKRSCR+RLA HN+RRRKT +
Sbjct: 190 GKRSCRKRLADHNRRRRKTQH 252
>AW691786 similar to PIR|T52298|T52 squamosa promoter binding protein-homolog
4 [imported] - garden snapdragon (fragment), partial
(42%)
Length = 649
Score = 119 bits (297), Expect = 8e-27
Identities = 56/90 (62%), Positives = 63/90 (69%)
Frame = +3
Query: 134 KKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQC 193
+K V +S CQVE C DL+ AK Y+ RHKVC MHSK+ V QRFCQQC
Sbjct: 171 RKEEVGQFNSSQPPRCQVEGCKLDLTDAKAYYSRHKVCSMHSKSPTVTVSGLQQRFCQQC 350
Query: 194 SRFHMLQEFDEGKRSCRRRLAGHNKRRRKT 223
SRFH L EFD+GKRSCRRRLAGHN+RRRKT
Sbjct: 351 SRFHQLAEFDQGKRSCRRRLAGHNERRRKT 440
>TC82440 similar to PIR|H96793|H96793 unknown protein F14G6.18 [imported] -
Arabidopsis thaliana, partial (12%)
Length = 1244
Score = 107 bits (266), Expect = 3e-23
Identities = 48/73 (65%), Positives = 59/73 (80%)
Frame = +1
Query: 134 KKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQC 193
K+ R +++ +CQV++C DLS+AKDYHRRHKVCE HSKA++AL+GN MQRFCQQC
Sbjct: 310 KRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQC 489
Query: 194 SRFHMLQEFDEGK 206
SRFH L EFDEGK
Sbjct: 490 SRFHPLVEFDEGK 528
Score = 42.4 bits (98), Expect = 0.001
Identities = 25/76 (32%), Positives = 36/76 (46%), Gaps = 7/76 (9%)
Frame = +2
Query: 155 CADLSRAKDYHRRHKVCEMHSKATRALV-------GNAMQRFCQQCSRFHMLQEFDEGKR 207
C L AK +++ ++ + + + +V G F F +G R
Sbjct: 353 CVKLITAKKIYQKRRIIIVVIRCAKLIVKLLRLC*GIKCNGFVNSVVGFIRWWSLMKGNR 532
Query: 208 SCRRRLAGHNKRRRKT 223
SCRRRLAGHN+RRRKT
Sbjct: 533 SCRRRLAGHNRRRRKT 580
>CA916954 similar to PIR|T52592|T525 squamosa-promoter binding protein 6
[imported] - Arabidopsis thaliana, partial (16%)
Length = 775
Score = 103 bits (256), Expect = 4e-22
Identities = 47/72 (65%), Positives = 53/72 (73%)
Frame = +3
Query: 144 SNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFD 203
S A CQV C DLS KDYH+RHKVCE+HSK +V QRFCQQCSRFH+L EFD
Sbjct: 558 SMTAYCQVYGCNKDLSSCKDYHKRHKVCEVHSKTPIVIVNGIEQRFCQQCSRFHLLSEFD 737
Query: 204 EGKRSCRRRLAG 215
+G RSCR+RLAG
Sbjct: 738 DG*RSCRKRLAG 773
>TC81138 similar to PIR|T52606|T52606 squamosa promoter binding protein 7
[imported] - Arabidopsis thaliana (fragment), partial
(32%)
Length = 743
Score = 100 bits (250), Expect = 2e-21
Identities = 53/110 (48%), Positives = 69/110 (62%), Gaps = 6/110 (5%)
Frame = +2
Query: 134 KKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQC 193
K++R A AS R CQV C D+S K YHRRH+VC + A ++ ++R+CQQC
Sbjct: 389 KRARTARAAASAR--CQVPGCEVDISELKGYHRRHRVCLRCANAATVVLDGDVKRYCQQC 562
Query: 194 SRFHMLQEFDEGKRSCRRRLAGHN-KRRRKTNNEAVPNGNPL-----NDD 237
+FH+L +FDEGKRSCRR+L HN +RRRK N AV N + NDD
Sbjct: 563 GKFHVLSDFDEGKRSCRRKLERHNTRRRRKAVNSAVGVDNEVQTVTQNDD 712
>BG582086 similar to PIR|T52603|T526 squamosa promoter binding protein 2
[imported] - Arabidopsis thaliana, partial (18%)
Length = 805
Score = 100 bits (249), Expect = 3e-21
Identities = 48/95 (50%), Positives = 62/95 (64%)
Frame = +1
Query: 134 KKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQC 193
KK + G CQVE C +LS AKDYH + +VCE H+K+ ++ +RFCQQC
Sbjct: 169 KKCKSNGQNLQCPPHCQVEGCGLNLSSAKDYHCKRRVCESHAKSPMVVIDGLERRFCQQC 348
Query: 194 SRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEAV 228
SRFH L EFD K+SCRR+L+ HN RRRK + +AV
Sbjct: 349 SRFHDLFEFDGKKKSCRRQLSNHNARRRKYHRQAV 453
>TC91858 similar to PIR|T52592|T52592 squamosa-promoter binding protein 6
[imported] - Arabidopsis thaliana, partial (19%)
Length = 1130
Score = 89.7 bits (221), Expect = 5e-18
Identities = 47/111 (42%), Positives = 60/111 (53%), Gaps = 12/111 (10%)
Frame = +3
Query: 106 NEEGGTLSLKLGGHAADRE-----------VASWDGGNGKKSRVAGGGASNR-AVCQVED 153
N + LKLG R+ + S G + RV ++ A CQV
Sbjct: 798 NGSNSLIDLKLGRFGDHRDGIGTPFSKGTTILSSSGSSTPSKRVRSSAIHSQIAYCQVYG 977
Query: 154 CCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDE 204
C DLS KDYH+RHKVCE+HSK ++ +V QRFCQQCSRFH+L EFD+
Sbjct: 978 CNKDLSSCKDYHKRHKVCEVHSKTSKVIVNGIEQRFCQQCSRFHLLAEFDD 1130
>TC78902
Length = 686
Score = 66.6 bits (161), Expect = 4e-11
Identities = 27/41 (65%), Positives = 31/41 (74%)
Frame = +1
Query: 1 MEARFGAEAYHYYGVGASSDMRGMGKRSAEWDLNNWRWDGD 41
M R GAE YH+YGVG SSD+ GMG S EW+LN+W WDGD
Sbjct: 562 MGERLGAENYHFYGVGGSSDLSGMGXXSREWNLNDWXWDGD 684
>TC92185 similar to GP|13374857|emb|CAC34491. putative protein {Arabidopsis
thaliana}, partial (5%)
Length = 557
Score = 63.2 bits (152), Expect = 5e-10
Identities = 33/61 (54%), Positives = 40/61 (65%), Gaps = 2/61 (3%)
Frame = +3
Query: 8 EAYHYYGVGA-SSDMRGMGKRSAEWDLNNWRWDGDLFIA-SRVNPVQEDGVGQQLFPLVS 65
EA+ YG G SSD+R MGK S EWDLNNW+WD LFIA S++ PV E +Q P+
Sbjct: 381 EAFQLYGNGGGSSDLRAMGKNSKEWDLNNWKWDSHLFIATSKLTPVPEH---RQFLPIPV 551
Query: 66 G 66
G
Sbjct: 552 G 554
>BG587629 similar to PIR|T52605|T526 squamosa promoter binding protein 7
[imported] - Arabidopsis thaliana, partial (9%)
Length = 631
Score = 57.8 bits (138), Expect = 2e-08
Identities = 45/171 (26%), Positives = 74/171 (42%), Gaps = 20/171 (11%)
Frame = +2
Query: 573 VSPIAAPASKRVQFSVKGVNMIRPATRLMCALEGKYLVCEDAHESMDQYSEELDGLHCI- 631
+ P A K ++F G N+++P RL+ + GKYL CE S +E D + C
Sbjct: 101 IHPTCFEAGKPMEFFACGSNLLQPKFRLLVSFYGKYLKCEYCAPSPHNSAE--DNISCAF 274
Query: 632 ---QFSCAVPVTN----GRGFIEIEDQGLSSSFFPFIVAEEDVCSDICGLEPLLELSETD 684
+ VP G FIE+E++ S+F P ++ ++++C+++ L+ L+ S
Sbjct: 275 DNQLYKICVPHIEENLLGPAFIEVENESGLSNFIPVLIGDKEICTELKILQQKLDASLLS 454
Query: 685 PDIEGTGKIKAKSQAMDFIH----------EIGWLLHG--SQLKSRMVHLS 723
S F+H +I WLL S+ RMV S
Sbjct: 455 KQFRSASGSSICSSCEAFVHIHTSSSDLLVDIAWLLKDPTSENFDRMVSAS 607
>AW684082 homologue to PIR|T47773|T47 hypothetical protein F24I3.210 -
Arabidopsis thaliana, partial (35%)
Length = 496
Score = 35.0 bits (79), Expect = 0.14
Identities = 41/124 (33%), Positives = 54/124 (43%), Gaps = 13/124 (10%)
Frame = +2
Query: 771 DLALSEMGLLHRAVRRNSKQLVELL-------------LTYVPENISNEVRPEGKALVEG 817
DL ++ + RA+ + +LV+L+ L Y EN S EV KAL+E
Sbjct: 29 DLEDQKIRRMRRALDSSDVELVKLMVMGEGLNLDEALALPYAVENCSREVV---KALLEL 199
Query: 818 EKQSFLFRPDAAGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPE 877
F AGP G TPLHIAA + S D++ L + A N R G TP
Sbjct: 200 GAADVNF---PAGPTGKTPLHIAA-EMVSPDMVAVLLDH------HADPNVRTVDGVTPL 349
Query: 878 DYAR 881
D R
Sbjct: 350 DILR 361
>AW559848 homologue to GP|21592490|gb| unknown {Arabidopsis thaliana},
partial (13%)
Length = 506
Score = 32.7 bits (73), Expect = 0.71
Identities = 22/70 (31%), Positives = 33/70 (46%), Gaps = 6/70 (8%)
Frame = +1
Query: 117 GGHAADREVASWDGGNGKKSRVAGG--GASNRAVCQVEDCCADLSRAKDY----HRRHKV 170
GG + + DGG+GK+S+ +G G N + E+ + KDY RR +
Sbjct: 265 GGARGNSNAVNDDGGDGKRSKTSGNSKGEENSSGKHAEETSDEPHPKKDYIHVRARRGQA 444
Query: 171 CEMHSKATRA 180
+ HS A RA
Sbjct: 445 TDSHSLAERA 474
>TC83350 similar to GP|21555074|gb|AAM63770.1 unknown {Arabidopsis
thaliana}, partial (54%)
Length = 582
Score = 31.6 bits (70), Expect = 1.6
Identities = 11/27 (40%), Positives = 16/27 (58%)
Frame = +3
Query: 21 MRGMGKRSAEWDLNNWRWDGDLFIASR 47
M R+ EWD+N W W+G L + S+
Sbjct: 144 MSAASYRADEWDVNKWAWEGILKVVSK 224
>TC86828 similar to PIR|T08454|T08454 hypothetical protein F22O6.170 -
Arabidopsis thaliana, partial (23%)
Length = 826
Score = 30.4 bits (67), Expect = 3.5
Identities = 17/62 (27%), Positives = 27/62 (43%), Gaps = 3/62 (4%)
Frame = +3
Query: 648 IEDQGLSSSFFPFIVAEEDVCSDICGLEPLLELSETDPDIEGTG---KIKAKSQAMDFIH 704
IE G + CS C L+E S D D+EG KI A+ +A++ ++
Sbjct: 465 IESAGAVEFLASIVTQNNTSCSSSCSATELIEASFDDDDVEGFAFDFKIGAEDEAINILY 644
Query: 705 EI 706
+
Sbjct: 645 NL 650
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,554,325
Number of Sequences: 36976
Number of extensions: 457895
Number of successful extensions: 2408
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 2360
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2392
length of query: 1003
length of database: 9,014,727
effective HSP length: 106
effective length of query: 897
effective length of database: 5,095,271
effective search space: 4570458087
effective search space used: 4570458087
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0158.4


