

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
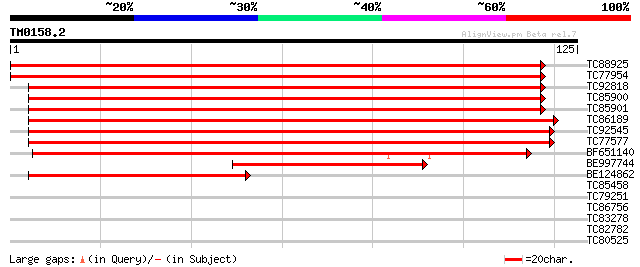
Query= TM0158.2
(125 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88925 similar to GP|19912165|dbj|BAB88394. autophagy 8h {Arabi... 210 1e-55
TC77954 similar to GP|19912167|dbj|BAB88395. autophagy 8i {Arabi... 180 1e-46
TC92818 homologue to GP|21592920|gb|AAM64870.1 putative microtub... 139 2e-34
TC85900 homologue to GP|21592920|gb|AAM64870.1 putative microtub... 138 6e-34
TC85901 similar to GP|20160828|dbj|BAB89768. contains ESTs C9802... 138 6e-34
TC86189 similar to GP|19912151|dbj|BAB88387. autophagy 8a {Arabi... 137 1e-33
TC92545 homologue to GP|18376155|emb|CAD21230. probable autophag... 131 6e-32
TC77577 GP|21615419|emb|CAD33929. microtubule associated protein... 125 4e-30
BF651140 weakly similar to GP|21592920|gb putative microtubule-a... 97 2e-21
BE997744 homologue to GP|15724332|gb| At2g45170/T14P1.2 {Arabido... 50 2e-07
BE124862 similar to GP|19912151|dbj autophagy 8a {Arabidopsis th... 50 3e-07
TC85458 weakly similar to GP|1817544|dbj|BAA11682.1 proline oxid... 26 4.6
TC79251 similar to GP|21555541|gb|AAM63881.1 unknown {Arabidopsi... 25 6.0
TC86756 weakly similar to GP|18253083|dbj|BAB83948. CIG1 {Nicoti... 25 6.0
TC83278 similar to GP|9294065|dbj|BAB02022.1 contains similarity... 25 6.0
TC82782 similar to GP|14532702|gb|AAK64152.1 unknown protein {Ar... 25 7.9
TC80525 similar to GP|15450715|gb|AAK96629.1 At2g32600/T26B15.16... 25 7.9
>TC88925 similar to GP|19912165|dbj|BAB88394. autophagy 8h {Arabidopsis
thaliana}, partial (86%)
Length = 739
Score = 210 bits (535), Expect = 1e-55
Identities = 101/118 (85%), Positives = 111/118 (93%)
Frame = +1
Query: 1 MGGSSSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVG 60
MGG S F++EFTF+QRLEESR+IIAKYPDR+PVIVE+Y K DLP L K+KYLVPRDLSVG
Sbjct: 76 MGGGSYFQNEFTFEQRLEESRDIIAKYPDRIPVIVEKYTKCDLPHLEKKKYLVPRDLSVG 255
Query: 61 HFIHILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFG 118
HFIHILSSRL+LP GKALFVFVKNTLPQTASMMDSVYRSF+DEDGFLY+YYSTEKTFG
Sbjct: 256 HFIHILSSRLNLPAGKALFVFVKNTLPQTASMMDSVYRSFKDEDGFLYLYYSTEKTFG 429
>TC77954 similar to GP|19912167|dbj|BAB88395. autophagy 8i {Arabidopsis
thaliana}, partial (98%)
Length = 1066
Score = 180 bits (457), Expect = 1e-46
Identities = 83/118 (70%), Positives = 104/118 (87%)
Frame = +1
Query: 1 MGGSSSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVG 60
MG +SFK EF+ D+R +ES+ II KYPDR+PVI+E+Y ++DLPEL K+KYLVPRD+SVG
Sbjct: 355 MGKINSFKQEFSLDERRKESQSIIVKYPDRIPVIIEKYSRTDLPELDKKKYLVPRDMSVG 534
Query: 61 HFIHILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFG 118
FIHILSSRL L PGKALF+FVKNTLPQTAS+M S+Y+++++EDGFLYM YS+EKTFG
Sbjct: 535 QFIHILSSRLRLTPGKALFIFVKNTLPQTASLMTSIYQTYKEEDGFLYMCYSSEKTFG 708
>TC92818 homologue to GP|21592920|gb|AAM64870.1 putative
microtubule-associated protein {Arabidopsis thaliana},
partial (97%)
Length = 656
Score = 139 bits (351), Expect = 2e-34
Identities = 65/114 (57%), Positives = 85/114 (74%)
Frame = +2
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
SSFK E ++R E+ I KYPDR+PVIVER K+D+PE+ K+KYLVP DL+VG F++
Sbjct: 143 SSFKTEHPLERRQAEAARIREKYPDRIPVIVERAEKTDVPEIDKKKYLVPADLTVGQFVY 322
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFG 118
++ R+ L P KA+F+FVKN LP TA+MM ++Y +DEDGFLYM YS E TFG
Sbjct: 323 VVRKRIKLSPEKAIFIFVKNILPPTAAMMSAIYEENKDEDGFLYMTYSGENTFG 484
>TC85900 homologue to GP|21592920|gb|AAM64870.1 putative
microtubule-associated protein {Arabidopsis thaliana},
partial (97%)
Length = 685
Score = 138 bits (347), Expect = 6e-34
Identities = 65/114 (57%), Positives = 84/114 (73%)
Frame = +1
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
SSFK E ++R E+ I KYPDR+PVIVER KSD+P++ K+KYLVP DL+VG F++
Sbjct: 130 SSFKLEHPLERRQAEAARIREKYPDRIPVIVERAEKSDVPDIDKKKYLVPADLTVGQFVY 309
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFG 118
++ R+ L P KA+F+FVKN LP TA MM ++Y +DEDGFLYM YS E TFG
Sbjct: 310 VVRKRIKLSPEKAIFIFVKNILPPTAGMMSAIYEENKDEDGFLYMTYSGENTFG 471
>TC85901 similar to GP|20160828|dbj|BAB89768. contains ESTs C98021(C0384)
D15269(C0384)~similar to Arabidopsis thaliana chromosome
3 At3g13230~, partial (88%)
Length = 1484
Score = 138 bits (347), Expect = 6e-34
Identities = 65/114 (57%), Positives = 84/114 (73%)
Frame = -1
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
SSFK E ++R E+ I KYPDR+PVIVER KSD+P++ K+KYLVP DL+VG F++
Sbjct: 389 SSFKLEHPLERRQAEAARIREKYPDRIPVIVERAEKSDVPDIDKKKYLVPADLTVGQFVY 210
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFG 118
++ R+ L P KA+F+FVKN LP TA MM ++Y +DEDGFLYM YS E TFG
Sbjct: 209 VVRKRIKLSPEKAIFIFVKNILPPTAGMMSAIYEENKDEDGFLYMTYSGENTFG 48
>TC86189 similar to GP|19912151|dbj|BAB88387. autophagy 8a {Arabidopsis
thaliana}, partial (96%)
Length = 735
Score = 137 bits (345), Expect = 1e-33
Identities = 64/117 (54%), Positives = 87/117 (73%)
Frame = +2
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
+SFK + F++R ES I KYPDRVPVIVE+ +SD+ ++ K+KYLVP DLSVG F++
Sbjct: 233 ASFKTQHPFERRQAESSRIREKYPDRVPVIVEKAGRSDIADIDKKKYLVPADLSVGQFVY 412
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGSVH 121
++ R+ L KA+FVF++NTLP TA++M ++Y +DEDGFLYM YS E TFGS H
Sbjct: 413 VVRKRIKLSAEKAIFVFIENTLPPTAALMSALYEEHKDEDGFLYMTYSGENTFGSSH 583
>TC92545 homologue to GP|18376155|emb|CAD21230. probable autophagy protein
AUT7 {Neurospora crassa}, partial (96%)
Length = 722
Score = 131 bits (330), Expect = 6e-32
Identities = 59/116 (50%), Positives = 82/116 (69%)
Frame = +2
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
S FKDE F++R E+ I KYPDR+PVI E+ KSD+ + K+KYLVP DL+VG F++
Sbjct: 59 SKFKDEHPFEKRKAEAERIRQKYPDRIPVICEKVEKSDIATIDKKKYLVPADLTVGQFVY 238
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGSV 120
++ R+ L P KA+F+FV LP TA++M S+Y +D+DGFLY+ YS E TFG +
Sbjct: 239 VIRKRIKLSPEKAIFIFVDEVLPPTAALMSSIYEEHKDDDGFLYITYSGENTFGDM 406
>TC77577 GP|21615419|emb|CAD33929. microtubule associated protein {Cicer
arietinum}, complete
Length = 992
Score = 125 bits (314), Expect = 4e-30
Identities = 57/116 (49%), Positives = 81/116 (69%)
Frame = +2
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
S FK E ++R E+ I KYPDR+PVIVE+ +SD+P + K+KYLVP DL+VG F++
Sbjct: 113 SYFKQEHDLEKRRAEAARIREKYPDRIPVIVEKAERSDIPSIDKKKYLVPADLTVGQFVY 292
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGSV 120
++ R+ L KA+F+FV N LP T ++M ++Y +DEDGFLY+ YS E TFG +
Sbjct: 293 VIRKRIKLSAEKAIFIFVDNVLPPTGAIMSAIYDEKKDEDGFLYVTYSGENTFGDL 460
>BF651140 weakly similar to GP|21592920|gb putative microtubule-associated
protein {Arabidopsis thaliana}, partial (83%)
Length = 645
Score = 97.1 bits (240), Expect = 2e-21
Identities = 52/112 (46%), Positives = 73/112 (64%), Gaps = 2/112 (1%)
Frame = +1
Query: 6 SFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIHI 65
S+K + +F++R E I K+ DRVPVIVE+ +SD+ ++ K+KYLVP DLSV F+ +
Sbjct: 142 SYKTKHSFERRQAEYNRIREKFSDRVPVIVEKAERSDIADIDKKKYLVPGDLSVAKFLFV 321
Query: 66 LSSRLSLPPGKALFVFV-KNTLPQTAS-MMDSVYRSFRDEDGFLYMYYSTEK 115
+ R+ L K +F FV N P AS +M S+Y +DEDGFLYM Y+ EK
Sbjct: 322 VRHRIRLSEDKDIFGFVHNNQSPVVASALMSSLYEEHKDEDGFLYMTYNEEK 477
>BE997744 homologue to GP|15724332|gb| At2g45170/T14P1.2 {Arabidopsis
thaliana}, partial (34%)
Length = 626
Score = 50.1 bits (118), Expect = 2e-07
Identities = 20/43 (46%), Positives = 30/43 (69%)
Frame = +1
Query: 50 KYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQTASM 92
+YLVP DL+VG F++++ R+ L KA+F+FV N LP T +
Sbjct: 439 RYLVPADLTVGQFVYVIRKRIKLSAEKAIFIFVDNVLPPTGKL 567
>BE124862 similar to GP|19912151|dbj autophagy 8a {Arabidopsis thaliana},
partial (40%)
Length = 675
Score = 49.7 bits (117), Expect = 3e-07
Identities = 24/49 (48%), Positives = 35/49 (70%)
Frame = +1
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLV 53
+SFK + F++R ES I KYPDRVPVIVE+ +SD+ ++ K+K +V
Sbjct: 91 ASFKTQHPFERRQAESSRIREKYPDRVPVIVEKAGRSDIADIDKKK*VV 237
>TC85458 weakly similar to GP|1817544|dbj|BAA11682.1 proline oxidase
precursor {Arabidopsis thaliana}, partial (69%)
Length = 1972
Score = 25.8 bits (55), Expect = 4.6
Identities = 13/39 (33%), Positives = 20/39 (50%)
Frame = +3
Query: 56 DLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQTASMMD 94
D ++ F+H + SLPP FV VK T S+++
Sbjct: 639 DRNLKGFLHTVDVSRSLPPSSVSFVIVKITAICPMSLLE 755
>TC79251 similar to GP|21555541|gb|AAM63881.1 unknown {Arabidopsis
thaliana}, partial (56%)
Length = 1253
Score = 25.4 bits (54), Expect = 6.0
Identities = 18/71 (25%), Positives = 30/71 (41%)
Frame = +2
Query: 42 DLPELGKRKYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFR 101
++P+ K L P +S LS+ + P G +F + LP + + SFR
Sbjct: 137 EIPKPSKPSSLNPVKVSSDLIFSALSTFTNFPAGFLSNLFDASRLPSNRNFSTNYIISFR 316
Query: 102 DEDGFLYMYYS 112
D + YY+
Sbjct: 317 DHTFAVPNYYT 349
>TC86756 weakly similar to GP|18253083|dbj|BAB83948. CIG1 {Nicotiana
tabacum}, partial (71%)
Length = 1715
Score = 25.4 bits (54), Expect = 6.0
Identities = 12/30 (40%), Positives = 16/30 (53%)
Frame = +2
Query: 56 DLSVGHFIHILSSRLSLPPGKALFVFVKNT 85
D ++ F+H + SLPP FV VK T
Sbjct: 581 DRNLKGFLHTVDVSKSLPPSSVSFVIVKIT 670
>TC83278 similar to GP|9294065|dbj|BAB02022.1 contains similarity to myb
proteins~gene_id:MRC8.8 {Arabidopsis thaliana}, partial
(11%)
Length = 920
Score = 25.4 bits (54), Expect = 6.0
Identities = 11/21 (52%), Positives = 14/21 (66%)
Frame = -3
Query: 51 YLVPRDLSVGHFIHILSSRLS 71
YLVPRD V F+H+ + LS
Sbjct: 858 YLVPRDKMVPSFLHVF*ASLS 796
>TC82782 similar to GP|14532702|gb|AAK64152.1 unknown protein {Arabidopsis
thaliana}, partial (46%)
Length = 760
Score = 25.0 bits (53), Expect = 7.9
Identities = 19/46 (41%), Positives = 24/46 (51%), Gaps = 3/46 (6%)
Frame = +3
Query: 30 RVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIHIL---SSRLSL 72
RVP I +L + L R +L+P D SVG F SSRLS+
Sbjct: 273 RVPTISFYFLGGPILTLPARNFLIPVD-SVGTFCFAFAPSSSRLSI 407
>TC80525 similar to GP|15450715|gb|AAK96629.1 At2g32600/T26B15.16
{Arabidopsis thaliana}, partial (63%)
Length = 591
Score = 25.0 bits (53), Expect = 7.9
Identities = 20/83 (24%), Positives = 37/83 (44%), Gaps = 4/83 (4%)
Frame = -1
Query: 39 LKSDLPELGKR----KYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQTASMMD 94
LK + LG R + +PR ++ F+H+ + L +++F F +T + M
Sbjct: 462 LKKERSLLGIRIKLLSHTIPRPPNLNRFLHVNLPLMRLRLSRSIFSFTSSTFSEIGLMAF 283
Query: 95 SVYRSFRDEDGFLYMYYSTEKTF 117
+ S + F+ + TE TF
Sbjct: 282 PL--SMSEVVTFVVVKSKTEFTF 220
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.140 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,445,214
Number of Sequences: 36976
Number of extensions: 40424
Number of successful extensions: 218
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 217
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 217
length of query: 125
length of database: 9,014,727
effective HSP length: 84
effective length of query: 41
effective length of database: 5,908,743
effective search space: 242258463
effective search space used: 242258463
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0158.2


