

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
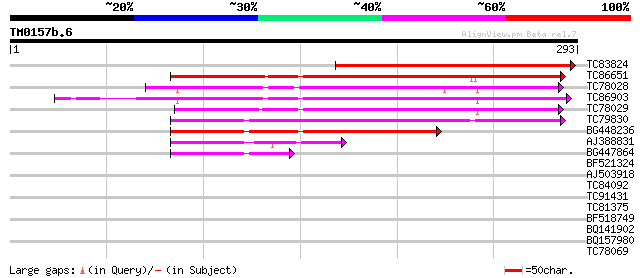
Query= TM0157b.6
(293 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83824 similar to GP|17381226|gb|AAL36425.1 unknown protein {Ar... 235 1e-62
TC86651 similar to PIR|F84555|F84555 similar to prolyl 4-hydroxy... 184 5e-47
TC78028 similar to GP|22136524|gb|AAM91340.1 unknown protein {Ar... 164 4e-41
TC86903 similar to GP|21617881|gb|AAM66931.1 prolyl 4-hydroxylas... 162 2e-40
TC78029 similar to GP|21593296|gb|AAM65245.1 prolyl 4-hydroxylas... 154 3e-38
TC79830 similar to PIR|G84861|G84861 hypothetical protein At2g43... 149 1e-36
BG448236 similar to GP|21537370|gb putative prolyl 4-hydroxylase... 110 6e-25
AJ388831 weakly similar to GP|10177121|dbj prolyl 4-hydroxylase ... 66 2e-11
BG447864 weakly similar to GP|10177121|db prolyl 4-hydroxylase ... 44 1e-04
BF521324 similar to GP|18086437|gb| AT3g28480/MFJ20_16 {Arabidop... 38 0.005
AJ503918 similar to GP|8131898|gb|A cyclic nucleotide-binding tr... 31 0.65
TC84092 weakly similar to GP|17644123|gb|AAL38986.1 cytochrome P... 29 1.9
TC91431 similar to GP|10177753|dbj|BAB11066. gene_id:MDN11.10~un... 29 2.5
TC81375 weakly similar to GP|21537367|gb|AAM61708.1 unknown {Ara... 28 5.5
BF518749 27 7.1
BQ141902 27 7.1
BQ157980 27 7.1
TC78069 27 9.3
>TC83824 similar to GP|17381226|gb|AAL36425.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 680
Score = 235 bits (600), Expect = 1e-62
Identities = 108/124 (87%), Positives = 116/124 (93%)
Frame = +3
Query: 169 HGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGL 228
HGEAFNILRYEV QRYN HYD+FNP EYGPQKSQR+ASFLLYLTDV+EGGETMFPFENGL
Sbjct: 3 HGEAFNILRYEVGQRYNSHYDAFNPDEYGPQKSQRVASFLLYLTDVEEGGETMFPFENGL 182
Query: 229 NMDVSYRYEDCIGLRVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWIRD 288
NMD +Y YEDC+GLRV+PRQGDGLLFYSL PNGTID TSLHGSCPVIKGEKWVATKWIR+
Sbjct: 183 NMDGTYGYEDCVGLRVKPRQGDGLLFYSLLPNGTIDQTSLHGSCPVIKGEKWVATKWIRN 362
Query: 289 HDQD 292
DQ+
Sbjct: 363 LDQE 374
>TC86651 similar to PIR|F84555|F84555 similar to prolyl 4-hydroxylase alpha
subunit [imported] - Arabidopsis thaliana, partial (85%)
Length = 1329
Score = 184 bits (466), Expect = 5e-47
Identities = 95/208 (45%), Positives = 134/208 (63%), Gaps = 4/208 (1%)
Frame = +2
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVS 143
+V+SW+PRA + NF T E+C+ ++ +AK + S + E ++++ +RTSSG F++
Sbjct: 353 EVVSWEPRAFVYHNFLTKEECEYLIDIAKPSMHKSTVVDSETGKSKDSR-VRTSSGTFLA 529
Query: 144 SSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQR 203
DK + IE+KIA T IP HGE +L YEV Q+Y PHYD F QR
Sbjct: 530 RGRDKI--VRNIEKKIADFTFIPVEHGEGLQVLHYEVGQKYEPHYDYFLDEFNTKNGGQR 703
Query: 204 MASFLLYLTDVQEGGETMFPFENGLNMDVSYRYE--DC--IGLRVRPRQGDGLLFYSLFP 259
+A+ L+YLTDV+EGGET+FP G +V + E DC GL ++P++GD LLF+S+ P
Sbjct: 704 IATVLMYLTDVEEGGETVFPAAKGNFSNVPWYNELSDCGKKGLSIKPKRGDALLFWSMKP 883
Query: 260 NGTIDPTSLHGSCPVIKGEKWVATKWIR 287
+ T+D +SLHG CPVIKG KW +TKWIR
Sbjct: 884 DATLDASSLHGGCPVIKGNKWSSTKWIR 967
>TC78028 similar to GP|22136524|gb|AAM91340.1 unknown protein {Arabidopsis
thaliana}, partial (86%)
Length = 1217
Score = 164 bits (415), Expect = 4e-41
Identities = 95/227 (41%), Positives = 137/227 (59%), Gaps = 11/227 (4%)
Frame = +1
Query: 71 GEFGDDSITSIPFQV--LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALR-EGET 127
G + S P +V +SWKPRA + F T +C ++ +AK+ LK SA+A GE+
Sbjct: 85 GSYAGTSAIIDPTKVKQVSWKPRAFVYKGFLTDLECDHLISIAKSELKRSAVADNLSGES 264
Query: 128 EQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPH 187
+ + +RTSSG+F+S ++D ++ IE+KI+ T +P+ +GE +LRYE Q+Y+PH
Sbjct: 265 KLSE--VRTSSGMFISKNKD--AIVSGIEDKISSWTFLPKENGEDIQVLRYEHGQKYDPH 432
Query: 188 YDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFP---FENGLNMDVSYRYEDCI---- 240
YD F + R+A+ L+YLT+V +GGET+FP + +S ED
Sbjct: 433 YDYFADKVNIARGGHRVATVLMYLTNVTKGGETVFPNAELQESPRHKLSETDEDLSECGK 612
Query: 241 -GLRVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWI 286
G+ V+PR+GD LLF+SL PN D SLH CPVI+GEKW ATKWI
Sbjct: 613 KGVAVKPRRGDALLFFSLHPNAIPDTLSLHAGCPVIEGEKWSATKWI 753
>TC86903 similar to GP|21617881|gb|AAM66931.1 prolyl 4-hydroxylase putative
{Arabidopsis thaliana}, partial (68%)
Length = 1310
Score = 162 bits (409), Expect = 2e-40
Identities = 100/273 (36%), Positives = 154/273 (55%), Gaps = 6/273 (2%)
Frame = +3
Query: 24 LICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESSEKAEYNLMTAGEFGDDSITSIPF 83
L+C FFF++ F S++ ++G N +T G + P
Sbjct: 183 LLCFFFFVS--FVSSIRLPGLEEG------------------NKITRGSVFGAKVKFDPT 302
Query: 84 QV--LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALRE-GETEQNTKGIRTSSGV 140
+V LSW PRA + NF T E+C ++ ++K L+ S +A E G++ Q+ +RTSSG+
Sbjct: 303 RVTQLSWSPRAFLYKNFLTDEECDHLIELSKDKLEKSMVADNESGKSIQSE--VRTSSGM 476
Query: 141 FVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQK 200
F++ +D+ ++ IE +IA T +P +GE+ +L Y ++Y PH+D F+
Sbjct: 477 FLNKQQDEI--VSGIEARIAAWTFLPVENGESMQVLHYMNGEKYEPHFDFFHDKANQRLG 650
Query: 201 SQRMASFLLYLTDVQEGGETMFPFENG-LNMDVSYRYEDCI--GLRVRPRQGDGLLFYSL 257
R+A+ L+YL++V++GGET+FP G L+ + +C G V+PR+GD LLF+SL
Sbjct: 651 GHRVATVLMYLSNVEKGGETIFPHAEGKLSQPKDESWSECAHKGYAVKPRKGDALLFFSL 830
Query: 258 FPNGTIDPTSLHGSCPVIKGEKWVATKWIRDHD 290
+ T D SLHGSCPVI+GEKW ATKWI D
Sbjct: 831 HLDATTDSKSLHGSCPVIEGEKWSATKWIHVAD 929
>TC78029 similar to GP|21593296|gb|AAM65245.1 prolyl 4-hydroxylase alpha
subunit-like protein {Arabidopsis thaliana}, partial
(89%)
Length = 1126
Score = 154 bits (390), Expect = 3e-38
Identities = 87/207 (42%), Positives = 124/207 (59%), Gaps = 6/207 (2%)
Frame = +3
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSS 145
+SW PRA + F T +C ++ +AK+ LK SA+A Q +RTSSG+F+S +
Sbjct: 228 ISWIPRAFVYQGFLTDLECDHLISLAKSELKRSAVADNLSGDSQ-LSDVRTSSGMFISKN 404
Query: 146 EDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMA 205
+D ++ IE++I+ T +P+ +GE +LRYE Q+Y+PHYD F Q R+A
Sbjct: 405 KDPI--VSGIEDRISAWTFLPKENGEDIQVLRYEHGQKYDPHYDYFADKVNIVQGGHRLA 578
Query: 206 SFLLYLTDVQEGGETMFP-FENGLNMDVSYRYEDCI-----GLRVRPRQGDGLLFYSLFP 259
+ L+YLT+V +GGET+FP E S + D G+ V+PR+GD LLF+SL
Sbjct: 579 TVLMYLTNVTKGGETVFPEAEEPPRRRGSKKSSDLSECAKKGIAVKPRRGDALLFFSLDT 758
Query: 260 NGTIDPTSLHGSCPVIKGEKWVATKWI 286
N D SLH CPV++GEKW ATKWI
Sbjct: 759 NAIPDTNSLHAGCPVLEGEKWSATKWI 839
>TC79830 similar to PIR|G84861|G84861 hypothetical protein At2g43080
[imported] - Arabidopsis thaliana, partial (87%)
Length = 1130
Score = 149 bits (377), Expect = 1e-36
Identities = 84/205 (40%), Positives = 120/205 (57%), Gaps = 1/205 (0%)
Frame = +1
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKG-IRTSSGVFV 142
+VLSW PR + NF + E+C + GVA LK S + + T + K +RTSSG+F+
Sbjct: 319 EVLSWSPRIILLHNFLSYEECDYLRGVALPRLKISTVV--DANTGKGIKSDVRTSSGMFL 492
Query: 143 SSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQ 202
S E K + IE++I+ + IP +GE +LRYE +Q Y PH+D F+ + Q
Sbjct: 493 SHEERKYPMIHAIEKRISVYSQIPIENGELMQVLRYEKNQYYRPHHDYFSDTFNLKRGGQ 672
Query: 203 RMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYSLFPNGT 262
R+A+ L+YL D EGGET FP + GL V+P +G+ +LF+S+ +G
Sbjct: 673 RIATMLMYLGDNVEGGETHFPSAGSDECSCGGKLTK--GLCVKPVKGNAVLFWSMGLDGQ 846
Query: 263 IDPTSLHGSCPVIKGEKWVATKWIR 287
DP S+HG CPV+ GEKW ATKW+R
Sbjct: 847 SDPDSVHGGCPVLAGEKWSATKWMR 921
>BG448236 similar to GP|21537370|gb putative prolyl 4-hydroxylase alpha
subunit {Arabidopsis thaliana}, partial (52%)
Length = 682
Score = 110 bits (275), Expect = 6e-25
Identities = 57/141 (40%), Positives = 90/141 (63%), Gaps = 1/141 (0%)
Frame = +3
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKG-IRTSSGVFV 142
++LSW+PRA + NF + E+C+ ++ +AK L S++ + +T ++T+ +RTSSG+F+
Sbjct: 204 EILSWEPRAFVYHNFLSKEECEHLINLAKPFLAKSSVV--DSKTGKSTESRVRTSSGMFL 377
Query: 143 SSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQ 202
+DK + IE +IA T IP +GE +L Y V ++Y PHYD F Q
Sbjct: 378 KRGKDKI--IQNIERRIADFTFIPVENGEGLQVLHYGVGEKYEPHYDYFLDEFNTKNGGQ 551
Query: 203 RMASFLLYLTDVQEGGETMFP 223
R+A+ L+YL+DV+EGGET+FP
Sbjct: 552 RVATVLMYLSDVEEGGETVFP 614
>AJ388831 weakly similar to GP|10177121|dbj prolyl 4-hydroxylase alpha
subunit-like protein {Arabidopsis thaliana}, partial
(33%)
Length = 505
Score = 65.9 bits (159), Expect = 2e-11
Identities = 36/95 (37%), Positives = 53/95 (54%), Gaps = 4/95 (4%)
Frame = +3
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGI----RTSSG 139
Q++SW+PRA + NF T E+C+ ++ +AK + SA+ E+ G+ RTSSG
Sbjct: 198 QIISWEPRAFLYHNFLTKEECEHLINIAKPSMHKSAVI-----DEETGNGVDSSERTSSG 362
Query: 140 VFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFN 174
F+ D+ + IE +IA T IP HGE FN
Sbjct: 363 AFLKRGSDR--IVKNIERRIADFTFIPXEHGENFN 461
>BG447864 weakly similar to GP|10177121|db prolyl 4-hydroxylase alpha
subunit-like protein {Arabidopsis thaliana}, partial
(50%)
Length = 639
Score = 43.5 bits (101), Expect = 1e-04
Identities = 20/65 (30%), Positives = 36/65 (54%), Gaps = 1/65 (1%)
Frame = +3
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGET-EQNTKGIRTSSGVFV 142
+V+SW+P A + NF T E+C+ ++ + K + S + + ET + +RT SG F+
Sbjct: 408 EVVSWEPXAFVYHNFLTKEECEYLIDIXKPSMHKSTVV--DSETGKSKDSXVRTXSGTFL 581
Query: 143 SSSED 147
+ D
Sbjct: 582 ARGRD 596
>BF521324 similar to GP|18086437|gb| AT3g28480/MFJ20_16 {Arabidopsis
thaliana}, partial (15%)
Length = 285
Score = 37.7 bits (86), Expect = 0.005
Identities = 22/63 (34%), Positives = 32/63 (49%), Gaps = 2/63 (3%)
Frame = +1
Query: 64 EYNLMTAGEFGDDSITSIPFQV--LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALA 121
E N +T G + P +V LSW PRA + NF T E+C ++ ++K L+ S A
Sbjct: 94 EGNKITRGSVFGAKVKFDPTRVTQLSWSPRAFLYNNFLTDEECDHLIELSKDNLEKSMAA 273
Query: 122 LRE 124
E
Sbjct: 274 DNE 282
>AJ503918 similar to GP|8131898|gb|A cyclic nucleotide-binding transporter 1
{Arabidopsis thaliana}, partial (5%)
Length = 454
Score = 30.8 bits (68), Expect = 0.65
Identities = 14/26 (53%), Positives = 17/26 (64%)
Frame = +1
Query: 7 KGNLSSRSNKLTFPYIFLICIFFFLA 32
KG+L R K+ FP FL C FFFL+
Sbjct: 271 KGSL*YRFKKVIFPLYFLNCFFFFLS 348
>TC84092 weakly similar to GP|17644123|gb|AAL38986.1 cytochrome P450-3 {Musa
acuminata}, partial (27%)
Length = 679
Score = 29.3 bits (64), Expect = 1.9
Identities = 14/27 (51%), Positives = 17/27 (62%), Gaps = 2/27 (7%)
Frame = -1
Query: 19 FPYIFLI--CIFFFLAGFFGSTLFSHS 43
FP I L C FF +AGFF S+ +HS
Sbjct: 388 FPTILLTSSCAFFIMAGFFTSSAIAHS 308
>TC91431 similar to GP|10177753|dbj|BAB11066. gene_id:MDN11.10~unknown
protein {Arabidopsis thaliana}, partial (86%)
Length = 1047
Score = 28.9 bits (63), Expect = 2.5
Identities = 17/45 (37%), Positives = 24/45 (52%)
Frame = +1
Query: 37 STLFSHSQDDGRGLRPRPRLLESSEKAEYNLMTAGEFGDDSITSI 81
STLF+H D L+P R ES +Y M AGE+ + ++ I
Sbjct: 913 STLFAHIASDAV-LKPLGRFAESPLAKKYPXMCAGEYVEQELSVI 1044
>TC81375 weakly similar to GP|21537367|gb|AAM61708.1 unknown {Arabidopsis
thaliana}, partial (30%)
Length = 797
Score = 27.7 bits (60), Expect = 5.5
Identities = 13/33 (39%), Positives = 18/33 (54%)
Frame = -1
Query: 10 LSSRSNKLTFPYIFLICIFFFLAGFFGSTLFSH 42
LSS+ +K P +FLIC F S LF++
Sbjct: 431 LSSKRHKKAVPSLFLICFSITSIAFIASPLFNN 333
>BF518749
Length = 416
Score = 27.3 bits (59), Expect = 7.1
Identities = 21/91 (23%), Positives = 39/91 (42%)
Frame = +1
Query: 22 IFLICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESSEKAEYNLMTAGEFGDDSITSI 81
I + + F L FF +L + S D R L +E L + + + S
Sbjct: 13 ISMSLLTFLLFTFFTLSLLTTSFSDSR-----KELRNKNENVLKQLRNSVYYSNRIDPSR 177
Query: 82 PFQVLSWKPRALYFPNFATAEQCKSIVGVAK 112
Q+ SW+PR + F + ++C ++ +A+
Sbjct: 178 VVQI-SWQPRVFLYKGFLSDKECDYLISLAQ 267
>BQ141902
Length = 1112
Score = 27.3 bits (59), Expect = 7.1
Identities = 12/26 (46%), Positives = 18/26 (69%), Gaps = 1/26 (3%)
Frame = -1
Query: 16 KLTFPYIF-LICIFFFLAGFFGSTLF 40
+L FP++F + CIFF +GF+ LF
Sbjct: 116 QLLFPFMF*MFCIFFGESGFYFGVLF 39
>BQ157980
Length = 946
Score = 27.3 bits (59), Expect = 7.1
Identities = 21/52 (40%), Positives = 23/52 (43%), Gaps = 8/52 (15%)
Frame = -3
Query: 23 FLICIFFFLAGFFGSTLFSHS--------QDDGRGLRPRPRLLESSEKAEYN 66
F+ICIFF L F +F HS Q L PRP ES K YN
Sbjct: 254 FMICIFFNLFDFL---IFIHSNSVFK*NQQTLINLLGPRPNKTESPSKLVYN 108
>TC78069
Length = 1150
Score = 26.9 bits (58), Expect = 9.3
Identities = 12/18 (66%), Positives = 12/18 (66%)
Frame = +1
Query: 18 TFPYIFLICIFFFLAGFF 35
TFP FLICIFF FF
Sbjct: 10 TFPSSFLICIFFVFF*FF 63
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,193,585
Number of Sequences: 36976
Number of extensions: 124852
Number of successful extensions: 771
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 753
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 755
length of query: 293
length of database: 9,014,727
effective HSP length: 95
effective length of query: 198
effective length of database: 5,502,007
effective search space: 1089397386
effective search space used: 1089397386
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0157b.6


