

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0157b.4
(77 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
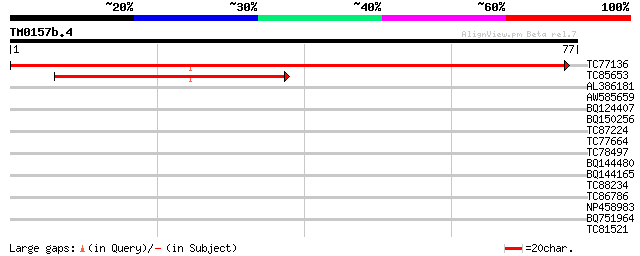
TC77136 homologue to GP|3860323|emb|CAA10129.1 hypothetical prot... 141 4e-35
TC85653 weakly similar to PIR|S44261|S44261 SRG1 protein - Arabi... 54 8e-09
AL386181 homologue to GP|3860323|emb|C hypothetical protein {Cic... 30 0.17
AW585659 similar to GP|2262110|gb|A zinc finger protein isolog {... 27 1.1
BQ124407 similar to GP|17979277|gb unknown protein {Arabidopsis ... 27 1.5
BQ150256 similar to GP|21166135|gb| hypothetical protein {Dictyo... 26 3.2
TC87224 weakly similar to GP|14532856|gb|AAK64110.1 unknown prot... 25 4.2
TC77664 similar to PIR|A86414|A86414 hypothetical protein AAF881... 25 4.2
TC78497 similar to PIR|T48092|T48092 hypothetical protein T20O10... 25 5.5
BQ144480 25 7.2
BQ144165 weakly similar to PIR|A49642|A496 transcription factor ... 25 7.2
TC88234 similar to GP|21592760|gb|AAM64709.1 unknown {Arabidopsi... 24 9.4
TC86786 similar to GP|8778315|gb|AAF79324.1| F14J16.29 {Arabidop... 24 9.4
NP458983 NP458983|AF498991.1|AAM18950.1 nodule-specific glycine-... 24 9.4
BQ751964 similar to GP|11994306|dbj emb|CAA66822.1~gene_id:MLJ15... 24 9.4
TC81521 24 9.4
>TC77136 homologue to GP|3860323|emb|CAA10129.1 hypothetical protein {Cicer
arietinum}, partial (97%)
Length = 584
Score = 141 bits (356), Expect = 4e-35
Identities = 68/77 (88%), Positives = 72/77 (93%), Gaps = 1/77 (1%)
Frame = +2
Query: 1 MGGGNAQKAKMARERNLEKQKGA-KGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHA 59
MGGGN QKAKMARERNLEKQK A KGSQLE NKKAMSIQCKVCMQTFICTTSEVKC+EHA
Sbjct: 89 MGGGNGQKAKMARERNLEKQKQAGKGSQLEKNKKAMSIQCKVCMQTFICTTSEVKCKEHA 268
Query: 60 EAKHPKSDVVTCFPHLQ 76
EA+HPKSD+ TCFPHL+
Sbjct: 269 EARHPKSDLFTCFPHLK 319
>TC85653 weakly similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis
thaliana, partial (20%)
Length = 660
Score = 54.3 bits (129), Expect = 8e-09
Identities = 30/33 (90%), Positives = 30/33 (90%), Gaps = 1/33 (3%)
Frame = +2
Query: 7 QKAKMARERNLEKQKGA-KGSQLETNKKAMSIQ 38
QKAKMARERNLEKQK A KGSQLE NKKAMSIQ
Sbjct: 80 QKAKMARERNLEKQKQAGKGSQLEKNKKAMSIQ 178
>AL386181 homologue to GP|3860323|emb|C hypothetical protein {Cicer
arietinum}, partial (37%)
Length = 208
Score = 30.0 bits (66), Expect = 0.17
Identities = 18/36 (50%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Frame = +2
Query: 3 GGNAQKAKMARERNLEKQKGA-KGSQLETNKKAMSI 37
GG K LEKQK A KGSQLE NKK++ +
Sbjct: 62 GGVRSKG*DGSREELEKQKQAGKGSQLEKNKKSVIV 169
>AW585659 similar to GP|2262110|gb|A zinc finger protein isolog {Arabidopsis
thaliana}, partial (27%)
Length = 483
Score = 27.3 bits (59), Expect = 1.1
Identities = 15/41 (36%), Positives = 24/41 (57%), Gaps = 1/41 (2%)
Frame = +3
Query: 24 KGSQLETNKKAMSIQCKVCM-QTFICTTSEVKCREHAEAKH 63
+GSQ NKK +++Q + + Q F+ TT+ V + AKH
Sbjct: 171 RGSQNNRNKKQLTVQGVIQLTQNFVTTTTIV*HNQDISAKH 293
>BQ124407 similar to GP|17979277|gb unknown protein {Arabidopsis thaliana},
partial (62%)
Length = 635
Score = 26.9 bits (58), Expect = 1.5
Identities = 14/43 (32%), Positives = 22/43 (50%), Gaps = 2/43 (4%)
Frame = +2
Query: 24 KGSQLETNKKAMSIQCKVCMQTFIC--TTSEVKCREHAEAKHP 64
K LE K++ I+C VC++ + T +E K +E HP
Sbjct: 125 KQKHLEALKRSQEIECSVCLERVLSKPTAAERKFGLLSECDHP 253
>BQ150256 similar to GP|21166135|gb| hypothetical protein {Dictyostelium
discoideum}, partial (4%)
Length = 303
Score = 25.8 bits (55), Expect = 3.2
Identities = 9/42 (21%), Positives = 24/42 (56%)
Frame = -2
Query: 6 AQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFI 47
A++ + R++ ++KG G LET+ ++S+ + + ++
Sbjct: 209 AKRRPVNRKKTQNREKGVLGPPLETSSLSLSLSLSIYLSLYL 84
>TC87224 weakly similar to GP|14532856|gb|AAK64110.1 unknown protein
{Arabidopsis thaliana}, partial (31%)
Length = 1760
Score = 25.4 bits (54), Expect = 4.2
Identities = 17/60 (28%), Positives = 26/60 (43%), Gaps = 8/60 (13%)
Frame = +3
Query: 21 KGAKG--SQLETNKKAMSIQCKVCMQTFICTT------SEVKCREHAEAKHPKSDVVTCF 72
KG+K + ETN A +Q + + F+C T S K +E E + + CF
Sbjct: 1263 KGSKDDHANAETNL*AFGLQIDILSEGFLCHTSYPTYISSDKAKEEEEQEQVMITFIVCF 1442
>TC77664 similar to PIR|A86414|A86414 hypothetical protein AAF88122.1
[imported] - Arabidopsis thaliana, partial (95%)
Length = 1597
Score = 25.4 bits (54), Expect = 4.2
Identities = 9/31 (29%), Positives = 16/31 (51%)
Frame = +2
Query: 30 TNKKAMSIQCKVCMQTFICTTSEVKCREHAE 60
+ K+ CK + ++C+TS +C AE
Sbjct: 650 SQSKSCPHSCKNSCKCYLCSTSSTRCHRFAE 742
>TC78497 similar to PIR|T48092|T48092 hypothetical protein T20O10.130 -
Arabidopsis thaliana, partial (56%)
Length = 911
Score = 25.0 bits (53), Expect = 5.5
Identities = 19/61 (31%), Positives = 27/61 (44%), Gaps = 10/61 (16%)
Frame = +2
Query: 13 RERNL--EKQKGAKGSQLETNKKAMS--------IQCKVCMQTFICTTSEVKCREHAEAK 62
RERNL E +K + + AM+ QCK+CM+ + T E E E +
Sbjct: 122 RERNLFAESMSASKTPSSSSKRPAMTQGSVDIYAAQCKLCMKWRVIDTQE----EFEEIR 289
Query: 63 H 63
H
Sbjct: 290 H 292
>BQ144480
Length = 1367
Score = 24.6 bits (52), Expect = 7.2
Identities = 10/41 (24%), Positives = 20/41 (48%)
Frame = -2
Query: 22 GAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAEAK 62
GA G + N + + C+V + + ++CRE A+ +
Sbjct: 1213 GAYGRGDDVNHREDRVLCRVKRYNIVLSDISIRCREKADTQ 1091
>BQ144165 weakly similar to PIR|A49642|A496 transcription factor Brn-3 - mouse,
partial (7%)
Length = 1309
Score = 24.6 bits (52), Expect = 7.2
Identities = 8/24 (33%), Positives = 15/24 (62%)
Frame = +1
Query: 30 TNKKAMSIQCKVCMQTFICTTSEV 53
T+ MS++C++C+Q C+ V
Sbjct: 1204 THSSMMSVRCRICLQHRRCSDMRV 1275
>TC88234 similar to GP|21592760|gb|AAM64709.1 unknown {Arabidopsis
thaliana}, partial (16%)
Length = 892
Score = 24.3 bits (51), Expect = 9.4
Identities = 14/47 (29%), Positives = 20/47 (41%)
Frame = +2
Query: 13 RERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHA 59
RER E++K +G T S C V +F TT+ H+
Sbjct: 14 RERKKERKKETRGDPQPTTLLNXSTFCSVLFFSFSLTTTTTITLTHS 154
>TC86786 similar to GP|8778315|gb|AAF79324.1| F14J16.29 {Arabidopsis
thaliana}, partial (32%)
Length = 707
Score = 24.3 bits (51), Expect = 9.4
Identities = 11/15 (73%), Positives = 12/15 (79%)
Frame = -1
Query: 26 SQLETNKKAMSIQCK 40
S L T+KKAMSI CK
Sbjct: 536 SNLITHKKAMSIACK 492
>NP458983 NP458983|AF498991.1|AAM18950.1 nodule-specific glycine-rich
protein 1B [Medicago truncatula]
Length = 393
Score = 24.3 bits (51), Expect = 9.4
Identities = 8/19 (42%), Positives = 12/19 (63%)
Frame = +3
Query: 41 VCMQTFICTTSEVKCREHA 59
VC+ +CTTS + C +A
Sbjct: 15 VCLCVLLCTTSYISCGNYA 71
>BQ751964 similar to GP|11994306|dbj emb|CAA66822.1~gene_id:MLJ15.12~unknown
protein {Arabidopsis thaliana}, partial (2%)
Length = 752
Score = 24.3 bits (51), Expect = 9.4
Identities = 11/34 (32%), Positives = 18/34 (52%)
Frame = -1
Query: 8 KAKMARERNLEKQKGAKGSQLETNKKAMSIQCKV 41
+A++ R+R L Q A GSQ +++ C V
Sbjct: 650 EAELGRQRRLRLQHCANGSQTRRQRRSRCRWCSV 549
>TC81521
Length = 869
Score = 24.3 bits (51), Expect = 9.4
Identities = 8/23 (34%), Positives = 14/23 (60%)
Frame = +2
Query: 32 KKAMSIQCKVCMQTFICTTSEVK 54
KK ++ ++C +TFIC + K
Sbjct: 791 KKIPTVNLRICYKTFICKFMQTK 859
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.125 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,468,084
Number of Sequences: 36976
Number of extensions: 28456
Number of successful extensions: 178
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 175
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 176
length of query: 77
length of database: 9,014,727
effective HSP length: 53
effective length of query: 24
effective length of database: 7,054,999
effective search space: 169319976
effective search space used: 169319976
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0157b.4


