

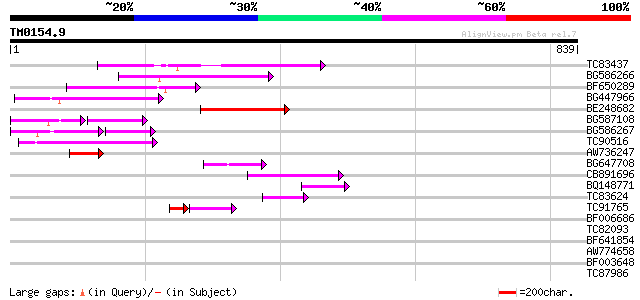
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0154.9
(839 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 192 3e-49
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 150 3e-36
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 147 2e-35
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 130 3e-30
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 127 2e-29
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 67 6e-25
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 80 2e-24
TC90516 105 7e-23
AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase... 70 4e-12
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 61 2e-09
CB891696 57 4e-08
BQ148771 46 7e-05
TC83624 homologue to PIR|G84581|G84581 copia-like retroelement p... 45 9e-05
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 37 1e-04
BF006686 37 0.031
TC82093 34 0.27
BF641854 similar to GP|6006861|gb|A putative auxin-independent g... 31 2.3
AW774658 similar to GP|2808681|emb| Hcr9-4B {Lycopersicon hirsut... 26 5.7
BF003648 similar to PIR|T01245|T01 N-acetyltransferase homolog F... 29 8.6
TC87986 similar to GP|5669636|gb|AAD46403.1| ethylene-responsive... 29 8.6
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 192 bits (489), Expect = 3e-49
Identities = 123/350 (35%), Positives = 195/350 (55%), Gaps = 12/350 (3%)
Frame = +2
Query: 130 RVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIVSKVLTTRLSKV 189
R + +FH N K NS+FI LIPKV+NP LND+ PISL+G +YKI+ K+L RL V
Sbjct: 5 RFVSEFHRNRKLFKGINSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVV 184
Query: 190 MGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKKRGFLVFKVDYEKVYDSVNWNF--- 246
+G +I + QSAF++ RQ+L+ V + + R ++ + + K++ + W
Sbjct: 185 IGSVISDAQSAFVKNRQILEMVFL*---------QMRLWMRLR-N*RKIFCCLRWILKRL 334
Query: 247 ------LFYMLRRLG--FYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGDPLA 298
L ++L +G F W WIK C+ +A+ SVLVNGSPT+
Sbjct: 335 ITLSIGLIWILF*VGMSFLVLWRKWIKECVSTATTSVLVNGSPTNV-------------- 472
Query: 299 PFLFLIVAECLSGMMRQARRLNLYKGFKVGREG-VEVSLLQFADDTLFIYEPSTHNVLAM 357
+M+ + L+ + G V VS LQFA+DTL + + N+ A+
Sbjct: 473 -------------LMKSLVQTQLFTRYSFGVVNPVVVSHLQFANDTLLLETKNWANIRAL 613
Query: 358 KSMLRCFELMSGLKVNFFKSKLAGVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVGANPR 417
++ L F MSGLKVNF KS L V++A L A++L K ++PF+YLG+ + N R
Sbjct: 614 RAALVIF*AMSGLKVNFHKSGLVCVNIAPSWLSEAASVLSWKVGKVPFLYLGMPIEGNSR 793
Query: 418 KKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKL 467
+ + W+P++++++ RL+ W + LSFGG++ L++SVL+S+ ++ L KL
Sbjct: 794 RLSFWEPIVNRIKARLTGWNSRFLSFGGRLVLLKSVLTSLSVYALPSSKL 943
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 150 bits (378), Expect = 3e-36
Identities = 79/232 (34%), Positives = 129/232 (55%), Gaps = 3/232 (1%)
Frame = -3
Query: 162 LNDYIPISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVID-- 219
+++Y I+ YKI++K+L+ R+ ++ II +QSAF+ GR + D+VL+ ++++
Sbjct: 778 VSEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYL 599
Query: 220 -EAKVKKRGFLVFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVN 278
++ KK + K D K YD + WNFL +L RLGF+ WI WI C+ + S S L+N
Sbjct: 598 RQSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLIN 419
Query: 279 GSPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQ 338
G P + L GDPL+P+LF++ E LSG+ +QA R G KV R ++ L
Sbjct: 418 GGPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARNCPPINHLL 239
Query: 339 FADDTLFIYEPSTHNVLAMKSMLRCFELMSGLKVNFFKSKLAGVSVAEDVLL 390
FADDT+F + + + + S++ + SG +N KS + S ++
Sbjct: 238 FADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAII 83
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 147 bits (370), Expect = 2e-35
Identities = 67/201 (33%), Positives = 120/201 (59%), Gaps = 3/201 (1%)
Frame = +3
Query: 85 LTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIKSFWHVLKDDIVRVLEDFHTNGVWPKE 144
L F E++ A++ + K+PG DGYN HF K W+++ D ++ + DF G PK
Sbjct: 12 LCSEFTAVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFMPKI 191
Query: 145 GNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAFLEG 204
N +++TL+PK N + ++ PI+ +YKI+SK+LT+R+ V+ ++ ENQSAF++G
Sbjct: 192 INCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVKG 371
Query: 205 RQLLDSVLVANEVIDEAKVKKRGF---LVFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWI 261
R + D++++++E++ ++G + K+D K YDS W F+ +++ LGF K++
Sbjct: 372 RVIFDNIILSHELV--KSYSRKGISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPYKFV 545
Query: 262 GWIKGCIQSASVSVLVNGSPT 282
W+ + +AS + NG T
Sbjct: 546 NWVMAXLTTASYTFNXNGDLT 608
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 130 bits (326), Expect = 3e-30
Identities = 75/225 (33%), Positives = 123/225 (54%), Gaps = 4/225 (1%)
Frame = +2
Query: 7 WVKDGDSNTKFFHNSINWRRRTNAIRGLVVD-GGWVEDPKVVKSKMKEFFEARFTNISGD 65
W+KDGD NTKFFH+ + RR+ N I+ L + G W + + V+ + +F FT S +
Sbjct: 5 WLKDGDKNTKFFHSKASQRRKVNEIKKLKDETGNWCKGEENVERLLITYFNNLFT--SSN 178
Query: 66 GVLLEGT---TFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIKSFWH 122
+E T +S E + + F EE+ A+ K+PGPDG F + +WH
Sbjct: 179 PTAIEETCEVVKGKLSHEHIVWCEKEFTEEEVLEAINQMHPVKAPGPDGLPALFFQKYWH 358
Query: 123 VLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIVSKVL 182
++ ++ +++ N + +E N +FI LIPK NP DY PISL + KI++KV+
Sbjct: 359 IVGKEVQQMVLQVLNNSMETEELNKTFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVI 538
Query: 183 TTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKKRG 227
R+ + + +ID QSAF++GR + D+ L+A V + +++G
Sbjct: 539 ANRVKQTLPDVIDVEQSAFVQGRLITDNALIAWSVSIG*RXRRKG 673
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 127 bits (319), Expect = 2e-29
Identities = 64/132 (48%), Positives = 90/132 (67%)
Frame = +3
Query: 283 DEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADD 342
+E +++ L GDPLAPFLFL+VAE +SG+M+ A NL++GF V R G VS LQ+ADD
Sbjct: 6 EEISVQRGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTRVSHLQYADD 185
Query: 343 TLFIYEPSTHNVLAMKSMLRCFELMSGLKVNFFKSKLAGVSVAEDVLLRYANLLHCKTTE 402
TL I P+ N+ +K++L+ FE+ SGLKVNF KS L G++V D + L+C+
Sbjct: 186 TLCIGMPTVDNLWTLKALLQGFEMASGLKVNFHKSSLIGINVPRDFMEAACRFLNCREES 365
Query: 403 IPFVYLGIHVGA 414
IPF+YLG+ G+
Sbjct: 366 IPFIYLGLPGGS 401
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 67.4 bits (163), Expect(2) = 6e-25
Identities = 34/89 (38%), Positives = 51/89 (57%)
Frame = +3
Query: 116 FIKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMY 175
F + WH++K D+++++ F +G N++ I LIPK P + + PISL Y
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 176 KIVSKVLTTRLSKVMGHIIDENQSAFLEG 204
KI+SKVL RL + +I E QSAF+ G
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFVHG 677
Score = 65.9 bits (159), Expect(2) = 6e-25
Identities = 43/115 (37%), Positives = 60/115 (51%), Gaps = 4/115 (3%)
Frame = +2
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLV-VDGGWVEDPKVVKSKMKEFFE--- 56
QKSR+ W GD N KF+H R N I GL DG W+ + + V+ ++FE
Sbjct: 59 QKSRNMWHISGDLNKKFYHALTKQRHARNRIVGLYDYDGNWITEEQGVEKVAVDYFEDLF 238
Query: 57 ARFTNISGDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDG 111
R T DG L E T+ S++ + N L R EE+R A++ +K+PGPDG
Sbjct: 239 QRTTPTGFDGFLDEITS--SITPQMNQRLLRLATEEEVRLALFIMHPEKAPGPDG 397
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 80.5 bits (197), Expect(2) = 2e-24
Identities = 51/146 (34%), Positives = 72/146 (48%), Gaps = 7/146 (4%)
Frame = +3
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGG---WVEDP--KVVKSKMKEFF 55
QKSR W++ GD NTKFFH RR N I L+ D +VE+ ++ S K +
Sbjct: 141 QKSRLNWLRSGDRNTKFFHAVTKNRRAQNRILSLIDDDDKEWFVEEDLGRLADSHFKLLY 320
Query: 56 EARFTNISGDGVLLE--GTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYN 113
+ G+ LE + V+EE N L + EE+R AV+D K PGPDG N
Sbjct: 321 SSEDV-----GITLEDWNSIPAIVTEEQNAQLMAQISREEVREAVFDINPHKCPGPDGMN 485
Query: 114 FHFIKSFWHVLKDDIVRVLEDFHTNG 139
F + FW + DD+ + ++F G
Sbjct: 486 VFFFQQFWDTMGDDLTSMAQEFLRTG 563
Score = 51.2 bits (121), Expect(2) = 2e-24
Identities = 29/73 (39%), Positives = 42/73 (56%)
Frame = +1
Query: 143 KEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAFL 202
+E L+PK L ++ PISL YKIVSKVL+ RL V+ II E Q+AF
Sbjct: 574 RESTKQTSGLVPKKLEAKRLVEFRPISLCNVAYKIVSKVLSKRLKSVLPWIITETQAAFG 753
Query: 203 EGRQLLDSVLVAN 215
+ + D++L+A+
Sbjct: 754 RRQLISDNILIAH 792
>TC90516
Length = 983
Score = 105 bits (262), Expect = 7e-23
Identities = 68/205 (33%), Positives = 109/205 (53%)
Frame = -2
Query: 14 NTKFFHNSINWRRRTNAIRGLVVDGGWVEDPKVVKSKMKEFFEARFTNISGDGVLLEGTT 73
NT +F + R N+I V+ W++D K + FF F++ +
Sbjct: 742 NTTYFLACVKNRGMRNSISAPRVER-WMDDVAKNKQAIVNFFTHHFSDP*TYRPTMGDID 566
Query: 74 FRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIKSFWHVLKDDIVRVLE 133
F +S DN+ L+ F + ++ V +G++SP PDG+N +F ++LK DI + E
Sbjct: 565 FFHISNLDNVLLSAQFLVSKMELVVSSLDGNESPRPDGFNLNFFIRLRNMLKADIEIMFE 386
Query: 134 DFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIVSKVLTTRLSKVMGHI 193
F+T K + F+TLI KV P+ L D+ +S +G + K+++KVL RL+ +M I
Sbjct: 385 QFYTPANLLKIFS*YFLTLISKVEYPILLGDFSLMSFLGSL*KLMAKVLALRLAHIMEKI 206
Query: 194 IDENQSAFLEGRQLLDSVLVANEVI 218
I NQS F+ GRQ +D V+ NE+I
Sbjct: 205 IFVNQSTFVRGRQHVDGVVAINEII 131
>AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (4%)
Length = 305
Score = 69.7 bits (169), Expect = 4e-12
Identities = 29/53 (54%), Positives = 39/53 (72%), Gaps = 2/53 (3%)
Frame = +1
Query: 89 FNLEEIRTAVWDCEGDKS--PGPDGYNFHFIKSFWHVLKDDIVRVLEDFHTNG 139
F+ EE+R AVWDC+ S PGPDG NF F+K +W ++K D +RV+ +FHTNG
Sbjct: 121 FSEEEVRKAVWDCDSSHS*NPGPDGVNFTFVKEYWELIKVDFLRVVMEFHTNG 279
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 61.2 bits (147), Expect = 2e-09
Identities = 34/94 (36%), Positives = 56/94 (59%), Gaps = 1/94 (1%)
Frame = +1
Query: 288 EKRLMLGDPLAPFLFLIVAECLSGMM-RQARRLNLYKGFKVGREGVEVSLLQFADDTLFI 346
EK L GDPL+P+LF++ A LSG++ R+ + NL+ G +V R +++ L FADD+L
Sbjct: 7 EKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLH-GIQVARSDPKITHLLFADDSLLF 183
Query: 347 YEPSTHNVLAMKSMLRCFELMSGLKVNFFKSKLA 380
+ + +L ++ SG VNF KS+++
Sbjct: 184 ARANLTEAATIMQVLHSYQSASGQLVNFEKSEVS 285
>CB891696
Length = 638
Score = 56.6 bits (135), Expect = 4e-08
Identities = 50/142 (35%), Positives = 69/142 (48%), Gaps = 1/142 (0%)
Frame = +1
Query: 353 NVLAMKSMLRCFELMSGLKVNFFKSKLAGVSVAEDVLLRYANLLHCKTTEIPFVYLGIHV 412
N+L MK+++ FEL S L VNF KS L ++V + CK + F YLGI V
Sbjct: 10 NILTMKTIVSYFELASSLWVNFLKSGLINLNVIGHF*GW*NIYIKCKVH*VIFKYLGILV 189
Query: 413 GANPRKKTTWDPLLSKLRKRL-SLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGV 471
G NP + + LL L L S W K L I + S + Y S K+P V
Sbjct: 190 GENPCRVNM*ELLLKLLTN*LGSWWNTK*LWTQNGFSQIHAK*ISQNI-YFSLMKIPVKV 366
Query: 472 ASLCNRIEKQFLWGGEEGTRKI 493
L ++++ QFL G + T+KI
Sbjct: 367 *ELISQLKTQFL*GNLKVTKKI 432
>BQ148771
Length = 680
Score = 45.8 bits (107), Expect = 7e-05
Identities = 20/71 (28%), Positives = 39/71 (54%)
Frame = -3
Query: 433 LSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVASLCNRIEKQFLWGGEEGTRK 492
L+ WK LS +V L +SV+ ++PL+ + +PK +++++F+WG E +R+
Sbjct: 573 LANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQKLQRKFVWGDTEVSRR 394
Query: 493 IAWVKWSKVCR 503
V W + +
Sbjct: 393 YHAVGWETMSK 361
>TC83624 homologue to PIR|G84581|G84581 copia-like retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 831
Score = 45.4 bits (106), Expect = 9e-05
Identities = 21/69 (30%), Positives = 37/69 (53%)
Frame = +1
Query: 374 FFKSKLAGVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVGANPRKKTTWDPLLSKLRKRL 433
F + +++ E + N L C E+PF +LG+ +GANP++ +T P+L L+ L
Sbjct: 256 FSRVNFMALNLEESFVEASPNFLLCNVNEVPFCFLGLPIGANPKRSSTRKPVLDSLQWLL 435
Query: 434 SLWKRKTLS 442
+ + T S
Sbjct: 436 YFFTKPTSS 462
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 37.4 bits (85), Expect(2) = 1e-04
Identities = 22/69 (31%), Positives = 33/69 (46%)
Frame = +2
Query: 267 CIQSASVSVLVNGSPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFK 326
C++S VLVN D + L GD L+P++F+I E LS ++ A+ G
Sbjct: 104 CVESNDYYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERGDTHGTS 283
Query: 327 VGREGVEVS 335
+ R VS
Sbjct: 284 I*RGAPPVS 310
Score = 26.9 bits (58), Expect(2) = 1e-04
Identities = 10/28 (35%), Positives = 20/28 (70%)
Frame = +1
Query: 237 KVYDSVNWNFLFYMLRRLGFYAKWIGWI 264
KVY+ V+ ++L ++ ++GF +WI W+
Sbjct: 13 KVYNRVD*DYLKEIMIKMGFNNRWIYWM 96
>BF006686
Length = 325
Score = 37.0 bits (84), Expect = 0.031
Identities = 16/40 (40%), Positives = 21/40 (52%)
Frame = +3
Query: 411 HVGANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLI 450
HV W+PLL + K L W K LSFGG++ L+
Sbjct: 204 HVEGRGEILPMWEPLLEHVNKMLKSWGNKLLSFGGRIVLL 323
>TC82093
Length = 569
Score = 33.9 bits (76), Expect = 0.27
Identities = 19/37 (51%), Positives = 24/37 (64%), Gaps = 1/37 (2%)
Frame = -1
Query: 2 KSRDRWVKDGDSNTKFFHN-SINWRRRTNAIRGLVVD 37
KS+ WVK G NT FFHN +N R +T+ GL+VD
Sbjct: 443 KSQ*NWVKYGSRNTWFFHNEDVN*RNQTS---GLIVD 342
>BF641854 similar to GP|6006861|gb|A putative auxin-independent growth
promoter protein {Arabidopsis thaliana}, partial (7%)
Length = 660
Score = 30.8 bits (68), Expect = 2.3
Identities = 13/37 (35%), Positives = 19/37 (51%)
Frame = +1
Query: 114 FHFIKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFI 150
FH KSFW + K I+ E + GV P + S++
Sbjct: 535 FHLCKSFWWICKHKIIEFQESLRSKGVSPMFKSFSYL 645
>AW774658 similar to GP|2808681|emb| Hcr9-4B {Lycopersicon hirsutum}, partial
(4%)
Length = 665
Score = 25.8 bits (55), Expect(2) = 5.7
Identities = 12/18 (66%), Positives = 15/18 (82%)
Frame = -3
Query: 356 AMKSMLRCFELMSGLKVN 373
A++S+L FE MSGLKVN
Sbjct: 324 ALRSILVIFENMSGLKVN 271
Score = 21.9 bits (45), Expect(2) = 5.7
Identities = 18/52 (34%), Positives = 26/52 (49%), Gaps = 1/52 (1%)
Frame = -1
Query: 310 SGMMRQARRLNLYKGFKVGREGVEV-SLLQFADDTLFIYEPSTHNVLAMKSM 360
S ++ + LNL +G + V S LQFADDTL + S N +S+
Sbjct: 449 SNLLSHSICLNL----SIGMHSLTVFSHLQFADDTLLLGVKSWANAPCGQSL 306
>BF003648 similar to PIR|T01245|T01 N-acetyltransferase homolog F16M14.6 -
Arabidopsis thaliana, partial (57%)
Length = 584
Score = 28.9 bits (63), Expect = 8.6
Identities = 15/52 (28%), Positives = 32/52 (60%), Gaps = 3/52 (5%)
Frame = -3
Query: 246 FLFYMLRRLGFYAKWI---GWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLG 294
FLF++ +R+ +A+W G I ++ + +L++ +PT+E + + L+LG
Sbjct: 579 FLFWLFKRVNVWARWFVKWGNIVALLEDTLLCLLLS-NPTEEEALPQNLLLG 427
>TC87986 similar to GP|5669636|gb|AAD46403.1| ethylene-responsive elongation
factor EF-Ts precursor {Lycopersicon esculentum},
partial (45%)
Length = 897
Score = 28.9 bits (63), Expect = 8.6
Identities = 22/75 (29%), Positives = 36/75 (47%), Gaps = 1/75 (1%)
Frame = -3
Query: 174 MYKIVSKVLTTRLSKVMGHIIDENQSAFLEGR-QLLDSVLVANEVIDEAKVKKRGFLVFK 232
M+KI +L +KV G + + QS E Q+L V+ NE+ + + GF+V
Sbjct: 643 MFKIQFHLL*RLRTKVKGRVAVQQQSLISERTCQMLKDVISGNEISLAIEFNESGFVVVL 464
Query: 233 VDYEKVYDSVNWNFL 247
+ E + S + FL
Sbjct: 463 SESE*TFRSGSRRFL 419
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.337 0.148 0.491
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,786,381
Number of Sequences: 36976
Number of extensions: 428158
Number of successful extensions: 3078
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1690
Number of HSP's successfully gapped in prelim test: 140
Number of HSP's that attempted gapping in prelim test: 1348
Number of HSP's gapped (non-prelim): 1895
length of query: 839
length of database: 9,014,727
effective HSP length: 104
effective length of query: 735
effective length of database: 5,169,223
effective search space: 3799378905
effective search space used: 3799378905
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0154.9


