

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0151.7
(1675 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
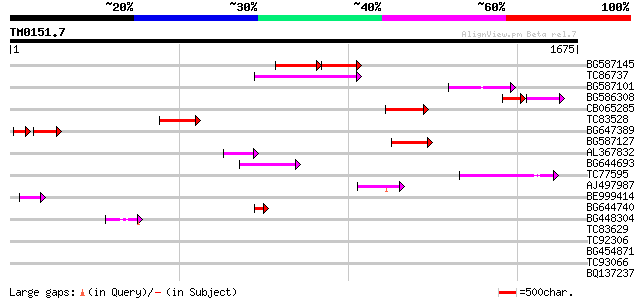
Score E
Sequences producing significant alignments: (bits) Value
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 169 1e-70
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 148 2e-35
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 119 8e-27
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 77 5e-26
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 99 2e-20
TC83528 95 2e-19
BG647389 69 1e-18
BG587127 weakly similar to PIR|H84506|H84 probable retroelement ... 91 4e-18
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 84 5e-16
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 79 1e-14
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 73 1e-12
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 67 8e-11
BE999414 59 2e-08
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 45 2e-04
BG448304 44 7e-04
TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNa... 42 0.002
TC92306 42 0.002
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 36 0.11
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 36 0.14
BQ137237 weakly similar to GP|11993889|gb| virion-associated nuc... 36 0.14
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 169 bits (428), Expect(2) = 1e-70
Identities = 77/136 (56%), Positives = 105/136 (76%)
Frame = +2
Query: 786 MHPADEDKTAFMTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVK 845
MHP D +KTAF+T R YCY+ MPFGLKNAG+TYQRL++R+FA ++G MEVY+DDM+VK
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 846 SVRGLDHHQDLEEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAIQ 905
S+R DH L+E F + ++ M+LNP KC+FGV G+FLG+++T +GIE+NP + AI
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 906 QMKNPSNVKEVQRLTG 921
+ +P N +EVQRLTG
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 117 bits (294), Expect(2) = 1e-70
Identities = 56/118 (47%), Positives = 81/118 (68%)
Frame = +3
Query: 921 GRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLKELLSSPPILSKPMQGH 980
GRIAAL+RF+ +S D+ PF+K L N F W +CEEAF +LK+ L++PP+LSKP G
Sbjct: 408 GRIAALNRFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGD 587
Query: 981 PLHLYFAVSDSALSSVILQEGDGEHRVIYFVSHTLQGAEVRYKKIEKAALAVLVTARR 1038
L LY A+S +A+SSV+++E GE + I++ S + E RY +EK A AV+ +AR+
Sbjct: 588 TLSLYIAISSTAVSSVLIREDRGEQKPIFYTSKRMTDPETRYPTLEKMAFAVITSARK 761
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 148 bits (373), Expect = 2e-35
Identities = 96/317 (30%), Positives = 151/317 (47%), Gaps = 3/317 (0%)
Frame = +1
Query: 724 ANVVMVKKANGKWLMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQ 783
A V+ V+K G CVDY LN KD YPLP I + +G + +D + +H+
Sbjct: 577 APVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLRRVAGARWFTKLDVVAAFHK 756
Query: 784 IRMHPADEDKTAFMTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMI 843
+R+ D++KTAF T + + PFGL A AT+QR +++ + + Y+DD++
Sbjct: 757 MRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYINKTLHEFLDDFVTAYIDDVL 936
Query: 844 VKSVRG-LDHHQDLEEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITS-RGIEINPDKC 901
+ + DH + + + L+P+KC F V K++GF++T+ +G+ +P K
Sbjct: 937 IYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVKYVGFILTAGKGVSCDPLKL 1116
Query: 902 KAIQQMKNPSNVKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFV 961
AI+ P +VK + G F+P + + P + +K+ F W AE E AF
Sbjct: 1117 AAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTRLTRKDFPFRWGAEQEAAFT 1296
Query: 962 RLKELLSSPPILSKPMQGHPLHLYFAVSDSALSSVILQE-GDGEHRVIYFVSHTLQGAEV 1020
+LK L + P+L + S AL V+ QE G G + F S L AE
Sbjct: 1297 KLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQEDGTGAAHPVAFHSQRLSPAEY 1476
Query: 1021 RYKKIEKAALAVLVTAR 1037
Y +K LAV R
Sbjct: 1477 NYPIHDKELLAVWACLR 1527
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 119 bits (299), Expect = 8e-27
Identities = 62/202 (30%), Positives = 105/202 (51%), Gaps = 3/202 (1%)
Frame = +2
Query: 1295 REASHYTLIDGHLYRRGFSAPLLKCVPPEKYEAIMSEVHEGMCASHIGGRSLACKVLRAG 1354
++ Y + LY++ ++CV E+ I+ H A H K+ +AG
Sbjct: 5 KDVRRYLWDEPFLYKQCADNIYIRCVAEEEIPGILFHCHGSNYAGHFAVSKTVSKIQQAG 184
Query: 1355 FYWPTLRKDCMDFVKKCKKCQVFADLS---KAPPKELVTMSAPWPFAMWGVDLVGPFPIA 1411
F+WPT+ KD F+ KC CQ ++S + P ++ + F +WG+D +GPFP +
Sbjct: 185 FWWPTMFKDAHSFISKCDPCQRQGNIS*RNEMPQNFILEVEV---FDVWGIDFMGPFPSS 355
Query: 1412 RSQMKFILVAVDYFTKWIEAEPLAKITSAKIVNFYWKRIVCRFGIPRAIVSDNGTQFSSN 1471
+ K+ILVAVDY +KW+EA + +V + I RFG+PR ++SD G+ F +
Sbjct: 356 YNN-KYILVAVDYVSKWVEAIASPTNDATVVVKMFKSVIFPRFGVPRVVISDGGSHFINK 532
Query: 1472 QTREFCREMGIQMRFASVEHPQ 1493
+ ++ G++ + A+ HPQ
Sbjct: 533 VFEKLLKKNGVRHKVATAYHPQ 598
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 76.6 bits (187), Expect(2) = 5e-26
Identities = 34/70 (48%), Positives = 49/70 (69%)
Frame = -2
Query: 1455 GIPRAIVSDNGTQFSSNQTREFCREMGIQMRFASVEHPQANGQVEYANRVILRGLRRRLK 1514
G+P IV+DNG+ F SN+ REFC I++ AS +PQ+NGQ E +N++I+ GL++RL
Sbjct: 682 GLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRLD 503
Query: 1515 EAKGAWLEEL 1524
KG W +EL
Sbjct: 502 LKKGCWADEL 473
Score = 61.2 bits (147), Expect(2) = 5e-26
Identities = 38/120 (31%), Positives = 64/120 (52%), Gaps = 7/120 (5%)
Frame = -3
Query: 1527 VLWSYNTTVQSTTRETPFRMTYGVDAMLPAEIDNFTW---RTEPDFEGENQANMAAELDL 1583
VLWS+ T + T+ TPF M + V+AM PAE++ + R + E N A L+
Sbjct: 465 VLWSHRTNPRGATKSTPFSMAHRVEAMAPAEVNVTSL*RSRMPQNIELNNDRLFNA-LET 289
Query: 1584 LSETRDEAHIRETAMKQRVAAKFNSRVRVRDMQVGDLVL----KWRSGASGNKLTPNWEG 1639
+ E RD+A +R + ++ + +N V+ + +++GD+VL + + KL NWEG
Sbjct: 288 IEERRDQALLRIQNYQHQIESYYNKTVKSQPLKLGDIVLCKVFENTKELNAGKLGTNWEG 109
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 98.6 bits (244), Expect = 2e-20
Identities = 53/128 (41%), Positives = 78/128 (60%)
Frame = -2
Query: 1110 DTQWTLFVDGSSNSSGSGTGVTLEGPGDLVLEQSLKFEFKATNNQAEYEALIAGLKLARE 1169
+++W L DG+ N+ G G G + P + + + F+ TNN AEYEA I G++ A +
Sbjct: 543 NSKWGLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNNMAEYEACIFGIEEAID 364
Query: 1170 VKIGSLLIRTDSQLVENQVKGTFQVKDPNLIKYLE*VRYLMTLFQEVVVEYVPRTENQRA 1229
++I L I DS LV NQ+KG ++ NLI Y + R L+T F +V + ++PR ENQ A
Sbjct: 363 MRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYFTKVELHHIPRDENQMA 184
Query: 1230 DALAKLAS 1237
DALA L+S
Sbjct: 183 DALATLSS 160
>TC83528
Length = 555
Score = 95.1 bits (235), Expect = 2e-19
Identities = 53/125 (42%), Positives = 80/125 (63%), Gaps = 4/125 (3%)
Frame = +3
Query: 442 PIVVQLRMNS-FNVRRVLLDQGSSADIIYGDAFDKLGLTDKDLTPDAGTLVGFAGEQVMV 500
P+V++L++N+ F+V RV ++ S DI+Y AF K+ L + L P G L G G+ + V
Sbjct: 180 PMVIKLQINNNFSVLRVFVNPMSKVDILYWSAFLKMKLQESMLKPCQGFLKGTFGKGLPV 359
Query: 501 RGYIDLDTIFGEDECARVLKVRYLVLQ---VVASYNVIIGRNTLNRLCAVISTAHLAVKY 557
+GYIDLDT FG+ E + +KVRY V++ V+ YNV++G +L L AV+S A +KY
Sbjct: 360 KGYIDLDTTFGKGENTKTIKVRYFVVESPPSVSIYNVVLGWPSLKDLKAVLSVAEFTIKY 539
Query: 558 PLSSG 562
P+ G
Sbjct: 540 PVGDG 554
>BG647389
Length = 718
Score = 68.6 bits (166), Expect(2) = 1e-18
Identities = 33/85 (38%), Positives = 54/85 (62%)
Frame = +1
Query: 69 KSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYMAR 128
K W+ LPR SI+ +++ + K + QFSA+K+ V+ +L+N+RQ ESL EY+
Sbjct: 451 KEATQQWYMNLPRFSITGYQNMTHKLVHQFSASKHCKVSTTNLFNVRQDPNESLPEYLV* 630
Query: 129 YSAASVKVEDEEPRACALAFKNGLL 153
++ A++KV + + A AF+NGLL
Sbjct: 631 FNNATIKVVNPKQELFAGAFQNGLL 705
Score = 44.7 bits (104), Expect(2) = 1e-18
Identities = 19/52 (36%), Positives = 33/52 (62%)
Frame = +2
Query: 10 RPFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAASDAVKC 61
+P S D+ + + +N+K+L L S+ SDP +H FNT+M +I A +++C
Sbjct: 278 QPLSMDIWNAPVLENLKSLSLLSFDDKSDPVEHATTFNTQMAVIGALKSLRC 433
>BG587127 weakly similar to PIR|H84506|H84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(13%)
Length = 415
Score = 90.9 bits (224), Expect = 4e-18
Identities = 48/119 (40%), Positives = 74/119 (61%)
Frame = +3
Query: 1129 GVTLEGPGDLVLEQSLKFEFKATNNQAEYEALIAGLKLAREVKIGSLLIRTDSQLVENQV 1188
G+ L P + +L+QS + EF A+NN+ YEALIAG++LA +KI ++ DSQLV +Q
Sbjct: 3 GIRLTSPTNEILKQSFRLEFHASNNETNYEALIAGVRLAHGLKIRNIHAYCDSQLVASQF 182
Query: 1189 KGTFQVKDPNLIKYLE*VRYLMTLFQEVVVEYVPRTENQRADALAKLASTRKPDNNRSV 1247
G ++ +D + YL+ V+ L + +PR+EN +ADALA LAS+ P+ R +
Sbjct: 183 SGEYEARDELMDTYLKLVQKLAQKLDYFALTRIPRSENVQADALAALASSSDPELKRVI 359
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 84.0 bits (206), Expect = 5e-16
Identities = 42/104 (40%), Positives = 61/104 (58%)
Frame = -1
Query: 631 RTLKIGTRLTEEQETRLTKLLGENLDLFAWSCKDMPGIDPNFICHRLALNPSVKPVSQLR 690
R +K+G L E + ++ +LL E LD+FA S +DMPG+DP + HR+ P PV
Sbjct: 312 REIKVGAALEEGVKRKIFQLLREYLDIFACSYEDMPGLDPKIVEHRIPTKPECPPVR*KL 133
Query: 691 RRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVMVKKANG 734
RR D ++ EV K + A F+ V+YP W+AN+V V K +G
Sbjct: 132 RRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDG 1
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 79.0 bits (193), Expect = 1e-14
Identities = 57/181 (31%), Positives = 86/181 (47%)
Frame = +2
Query: 679 LNPSVKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVMVKKANGKWLM 738
L P++ P+ R+ K K ++ ++ LL FI+ YP + V+ +KK +G M
Sbjct: 53 LLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVV-VLFLKKKDGFLRM 229
Query: 739 CVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQIRMHPADEDKTAFMT 798
+DY LN K YPLP ID L D G++ +D G HQ R+ D KTAF
Sbjct: 230 SIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVPKTAFRI 409
Query: 799 ARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEE 858
+Y M FG N + LM+RVF + + V+ +D+++ S +H L
Sbjct: 410 RYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEHENHLRL 589
Query: 859 A 859
A
Sbjct: 590 A 592
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 72.8 bits (177), Expect = 1e-12
Identities = 77/302 (25%), Positives = 125/302 (40%), Gaps = 8/302 (2%)
Frame = +2
Query: 1328 IMSEVHEGMCASHIGGRSLACKVLRAGFYWPTLRKDCMDFVKKCKKCQVFADLSKAPPKE 1387
++ E H+ A H GR+ +++ F+WP + FV+ C C +A
Sbjct: 179 LVQESHDSTAAGH-PGRNGTLEIVSRKFFWPGQSQTVRRFVRNCDVCGGIHIWRQAKRGF 355
Query: 1388 LVTMSAPWPF-AMWGVDLVGPFPIARSQ-MKFILVAVDYFTKWIEAEPLAKITSAKIVNF 1445
L + P + +D + P R + +++ V VD +K + E + + +
Sbjct: 356 LKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSKSVTLEEMDTMEAEACAQR 535
Query: 1446 YWKRIVCRFGIPRAIVSDNGTQFSSNQTREFCREMGIQMRFASVEHPQANGQVEYANRVI 1505
+ G+P++IVSD G+ + REFCR G+ ++ HPQ +G E N+ I
Sbjct: 536 FLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGTERWNQEI 715
Query: 1506 LRGLRRRLKEAKGAWLEELPVVLWSYNTTVQSTTRETPFRMTYGVDAMLPAEIDNFTWRT 1565
LR + ++ W + LP V + S+ TPF + +G +D T
Sbjct: 716 QAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGY------HVDPIP--T 871
Query: 1566 EPDFEGENQANMAAELDLLSETRD-----EAHIRETAMKQRVAAKFNSRVRVRD-MQVGD 1619
D G AA L+ +D +A I A +QR A N R D QVGD
Sbjct: 872 VEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAEI--VAAQQRSEASANKRRCPADRYQVGD 1045
Query: 1620 LV 1621
V
Sbjct: 1046 KV 1051
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 66.6 bits (161), Expect = 8e-11
Identities = 45/167 (26%), Positives = 74/167 (43%), Gaps = 29/167 (17%)
Frame = -2
Query: 1027 KAALAVLVTARRLRPYFQSFPVRVRTDL-PLRQVLQKPDMSGRLVAWSVELSEYGLQYDK 1085
K A+ A+RLR Y + + + + P++ + +KP ++GR+ W + LSEY ++Y
Sbjct: 635 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS 456
Query: 1086 RGKVGAQSLADFVV-----ELTPDRFERVDTQ-----------------------WTLFV 1117
+ + LAD + + P +F+ D + W L
Sbjct: 455 QKAIKGSILADHLAHQPLEDYRPIKFDFPDEEIMYLKMKDCDEPLFGEGPDPDSVWGLIF 276
Query: 1118 DGSSNSSGSGTGVTLEGPGDLVLEQSLKFEFKATNNQAEYEALIAGL 1164
DG+ N G+G G L P + + + F TNN AEYEA I G+
Sbjct: 275 DGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGI 135
>BE999414
Length = 613
Score = 58.9 bits (141), Expect = 2e-08
Identities = 26/76 (34%), Positives = 41/76 (53%)
Frame = +3
Query: 30 LDSYSGDSDPKDHLLYFNTKMVIIAASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRD 89
+DSY G DP +H+ + + AVKC++F +T + AM WF L R SI ++ D
Sbjct: 9 MDSYDGT*DPDEHMENIEVVLTYRSVRGAVKCKLFVTTLRRGAMTWFKNLRRNSIGSWGD 188
Query: 90 FSSKFLVQFSANKNQP 105
+F F+ ++ QP
Sbjct: 189 LCHEFTTHFTVSRTQP 236
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 45.4 bits (106), Expect = 2e-04
Identities = 19/41 (46%), Positives = 27/41 (65%)
Frame = -1
Query: 724 ANVVMVKKANGKWLMCVDYTDLNKACPKDSYPLPSIDSLVD 764
A ++ V+K +G + MC+DY NK K+ YPLP ID+L D
Sbjct: 238 AALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFD 116
>BG448304
Length = 637
Score = 43.5 bits (101), Expect = 7e-04
Identities = 33/117 (28%), Positives = 52/117 (44%), Gaps = 8/117 (6%)
Frame = +2
Query: 284 PEAPKYQSRDANPKKWCEFHRSAGHGTDDCWTLQREIDKLIRAGYQGNRQGQWRNNGDHN 343
P+ P +++ + K+C FH+ H TDDC L+ I+ LI+ G R Q N +
Sbjct: 65 PKTPTQENKGTDKTKYCRFHKCHRHLTDDCIHLKDTIEILIQRG----RLNQLTKNPEPE 232
Query: 344 KTHKREEERVDTKGKKKQESAAIATRGADDTFAQH--------SGPPVGTINTIAGG 392
K + ++ T K K A++ D+ F +H + P T N I GG
Sbjct: 233 K----QTVKLITDKKNKDIVVAMSVEQLDE-FVEHVDITPYSCTWEPFPTANVITGG 388
>TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNase H
{Arabidopsis thaliana}, partial (21%)
Length = 1071
Score = 42.0 bits (97), Expect = 0.002
Identities = 32/103 (31%), Positives = 49/103 (47%), Gaps = 7/103 (6%)
Frame = +2
Query: 1134 GPGDLVLEQ--SLKFEFKA-----TNNQAEYEALIAGLKLAREVKIGSLLIRTDSQLVEN 1186
G G L+L + SL + F+ T AEY AL+ GLK A + + DS+LV N
Sbjct: 602 GAGALLLAEDGSLLYGFRQGLGHQTKESAEYRALLLGLKHASMKGFKYVTAKGDSELVIN 781
Query: 1187 QVKGTFQVKDPNLIKYLE*VRYLMTLFQEVVVEYVPRTENQRA 1229
Q+ +++KD +L K L F ++++ R N A
Sbjct: 782 QILDPWKIKDEHLKKLCAEALELSDNFHSFRIQHISRERNYGA 910
>TC92306
Length = 521
Score = 42.0 bits (97), Expect = 0.002
Identities = 28/73 (38%), Positives = 37/73 (50%)
Frame = +2
Query: 1161 IAGLKLAREVKIGSLLIRTDSQLVENQVKGTFQVKDPNLIKYLE*VRYLMTLFQEVVVEY 1220
I GL AR + +R D Q V Q G+++V +PNL L + F+ V VE+
Sbjct: 2 ILGLNEARNQGYEHVHVRGDFQXVCKQFXGSWKVNNPNLRNLCNXAVELKSNFKSVSVEH 181
Query: 1221 VPRTENQRADALA 1233
VPR N ADA A
Sbjct: 182 VPRGXNXAADAQA 220
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 36.2 bits (82), Expect = 0.11
Identities = 23/82 (28%), Positives = 37/82 (45%)
Frame = +2
Query: 1470 SNQTREFCREMGIQMRFASVEHPQANGQVEYANRVILRGLRRRLKEAKGAWLEELPVVLW 1529
SN ++ + G + +S HP ++GQ E N+ LR + W + P +
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 1530 SYNTTVQSTTRETPFRMTYGVD 1551
YNT+ + TPF+ YG D
Sbjct: 212 WYNTSYNISAAMTPFKALYGRD 277
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 35.8 bits (81), Expect = 0.14
Identities = 21/70 (30%), Positives = 31/70 (44%), Gaps = 5/70 (7%)
Frame = +1
Query: 1458 RAIVSDNGTQFSSNQTREFCREMGIQMRFASVEHPQANGQVEYANRVILRGLRRRLKEA- 1516
+ +++DN +F S+ EFC GI +PQ NG E R +L R L A
Sbjct: 289 KKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAG 468
Query: 1517 ----KGAWLE 1522
+ W+E
Sbjct: 469 L*N*RDLWVE 498
>BQ137237 weakly similar to GP|11993889|gb| virion-associated
nuclear-shuttling protein {Mus musculus}, partial (4%)
Length = 1215
Score = 35.8 bits (81), Expect = 0.14
Identities = 19/66 (28%), Positives = 33/66 (49%)
Frame = +2
Query: 190 RKRAKLEKGDTSPKRAKKDKNGEDKGDGKQQRQDKGKAALRPTKEQLYPRRDDYEQRRPW 249
R R + +KG+ + + ++D N + Q++ KAA + ++ +RDD E RR
Sbjct: 833 RTRRQTKKGEAAERARRRDDNRTHSHTHRAPTQNQTKAAEQRPSNTVHKKRDDRENRRRM 1012
Query: 250 QSKSHR 255
Q S R
Sbjct: 1013QQASTR 1030
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,202,690
Number of Sequences: 36976
Number of extensions: 601334
Number of successful extensions: 2925
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 2863
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2914
length of query: 1675
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1566
effective length of database: 4,984,343
effective search space: 7805481138
effective search space used: 7805481138
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0151.7


