

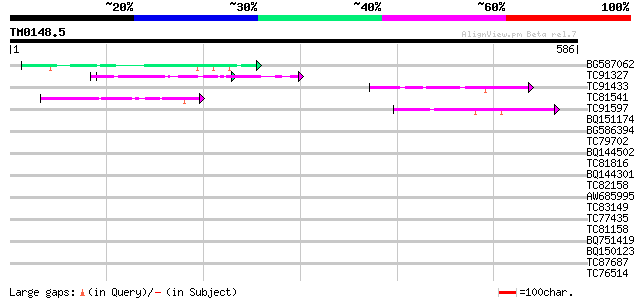
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0148.5
(586 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG587062 similar to GP|19881001|gb chemiosmotic efflux system B ... 58 9e-09
TC91327 similar to GP|22597168|gb|AAN03471.1 unknown protein {Gl... 53 3e-07
TC91433 50 2e-06
TC81541 similar to GP|22597168|gb|AAN03471.1 unknown protein {Gl... 44 2e-04
TC91597 similar to GP|21323350|dbj|BAB97978. Hypothetical protei... 42 7e-04
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 40 0.002
BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophi... 40 0.002
TC79702 MtN14 39 0.006
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 39 0.007
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 38 0.010
BQ144301 similar to GP|15277263|db alternative name: G2~unknown ... 37 0.021
TC82158 weakly similar to GP|19920013|gb|AAM08453.1 hypothetical... 37 0.028
AW685995 similar to PIR|I51618|I516 nucleolar phosphoprotein - A... 37 0.028
TC83149 similar to GP|13561982|gb|AAK30594.1 flagelliform silk p... 37 0.028
TC77435 similar to GP|7211427|gb|AAF40306.1| RNA helicase {Vigna... 36 0.047
TC81158 similar to GP|9280664|gb|AAF86533.1| F21B7.15 {Arabidops... 36 0.047
BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania maj... 35 0.081
BQ150123 similar to GP|21109992|gb| TonB-dependent receptor {Xan... 35 0.081
TC87687 similar to GP|8894548|emb|CAB95829.1 hypothetical protei... 35 0.081
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 35 0.11
>BG587062 similar to GP|19881001|gb chemiosmotic efflux system B protein A
{Legionella pneumophila}, partial (1%)
Length = 780
Score = 58.2 bits (139), Expect = 9e-09
Identities = 70/274 (25%), Positives = 102/274 (36%), Gaps = 26/274 (9%)
Frame = +3
Query: 13 KKRRREEKPEPLPSPEQDQEQSCSKKKR------RKKKKIKEEGIESHEHDQVLLPEPVV 66
+K REEKPE SPEQ+ E SKKK+ RKKKKI E P P
Sbjct: 39 QKMLREEKPEAFASPEQNPEDGSSKKKKKKRCRWRKKKKINEN------------PNPQP 182
Query: 67 VATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKET 126
+ P+ ++ P + E+ +PD T +K+KK
Sbjct: 183 QVCSEPLVVDAPISELKSNEESVT---LPDSNNIATKKKRKKNR---------------- 305
Query: 127 SNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKP-KEPCSTE 185
+ K K AE + EL PA++ G EPKP E E
Sbjct: 306 ------------NKNKNKSMINAEEPKPALELGKNEEPKPALELGKNEEPKPVLELEKNE 449
Query: 186 HPRVCPE-------PPLGLAIPIDPEKNRKE----AEPNESNAEHPRACPE--------P 226
P+ E + +A + ++ RK AE NE N E + P
Sbjct: 450 EPKPALELEKNKEVESIAVAKTMTRKRKRKSVQNGAESNEHNIEQAETAVQMSILDTEVP 629
Query: 227 PLGLAIPIDPEKNKGENLTPTDENELDRRNRREE 260
P+ AI ++ N+ +L +E +R+ R+E
Sbjct: 630 PIDKAIAVN---NQHVSLDELEERYFERKKTRKE 722
>TC91327 similar to GP|22597168|gb|AAN03471.1 unknown protein {Glycine max},
partial (70%)
Length = 738
Score = 53.1 bits (126), Expect = 3e-07
Identities = 58/220 (26%), Positives = 90/220 (40%)
Frame = +1
Query: 84 EPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKK 143
E ++ T++ D + AK ++K KK VL P+ + + N + CS +K
Sbjct: 52 EKVKVTIMKLKVDLQCAKCYKKVKK---VLCKFPQIRDQVYDEKNNIVTITVVCCSPEKI 222
Query: 144 KKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDP 203
+ + + A+ + V EPP P PKPKEP + P EP P +P
Sbjct: 223 RDKICCKGCGAIKSIEIV--EPP-------PPPKPKEPEKPKEPVKPKEPEK----PKEP 363
Query: 204 EKNRKEAEPNESNAEHPRACPEPPLGLAIPIDPEKNKGENLTPTDENELDRRNRREELVA 263
EK + +P E E P+ PE P P +PEK K + P E + + + E+
Sbjct: 364 EKPKNPEKPKEP--EKPKE-PEKPKEPEKPKEPEKPKEKPAPPPPEPKPEPPKQPEK--- 525
Query: 264 LVTSDEKIVNPRDHLVPLKSIPGFVLEQQTQPGPVIPVCP 303
P++ P + +P P IPVCP
Sbjct: 526 ----------PKEKPAPPP-------QPMPEPAPYIPVCP 594
Score = 46.6 bits (109), Expect = 3e-05
Identities = 40/145 (27%), Positives = 58/145 (39%)
Frame = +1
Query: 90 VVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGA 149
+V P P P K +K KE V +PE EP++ N + +P + K+ +K K
Sbjct: 271 IVEPPP---PPKPKEPEKPKEPVKPKEPEKPKEPEKPKNPE--KPKEPEKPKEPEKPKEP 435
Query: 150 ESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKE 209
E + + PP PEPKP+ P E P+ P PP
Sbjct: 436 EKPKEPEKPKEKPAPPP-------PEPKPEPPKQPEKPKEKPAPP--------------- 549
Query: 210 AEPNESNAEHPRACPEPPLGLAIPI 234
+P A + CP P +A+PI
Sbjct: 550 PQPMPEPAPYIPVCPPP---MAVPI 615
Score = 36.6 bits (83), Expect = 0.028
Identities = 43/160 (26%), Positives = 60/160 (36%), Gaps = 3/160 (1%)
Frame = +1
Query: 20 KPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITLETPK 79
+P P P P++ ++ K + +K + E ++ E + EP E PK
Sbjct: 277 EPPPPPKPKEPEKPKEPVKPKEPEKPKEPEKPKNPEKPK----EP-----------EKPK 411
Query: 80 EQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCS 139
E P + P K +K KE P PEP+PEP P Q
Sbjct: 412 E------------PEKPKEPEKPKEPEKPKEKPAPPPPEPKPEP----------PKQ--- 516
Query: 140 VKKKKKRKGAESNEAVAELA---TVCTEPPAVQGGVLPEP 176
+K K K A + + E A VC P AV GV P
Sbjct: 517 -PEKPKEKPAPPPQPMPEPAPYIPVCPPPMAVPIGVCCTP 633
Score = 30.8 bits (68), Expect = 1.5
Identities = 25/92 (27%), Positives = 41/92 (44%)
Frame = +1
Query: 8 ELLHSKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVV 67
E + K+ + ++PE +PE+ +E K K +K K E+ E + + P P
Sbjct: 322 EPVKPKEPEKPKEPEKPKNPEKPKEPE--KPKEPEKPKEPEKPKEPEKPKEKPAPPPPEP 495
Query: 68 ATAIPITLETPKEQGAEPIEATVVHPVPDQRP 99
P E PKE+ A P + P+P+ P
Sbjct: 496 KPEPPKQPEKPKEKPAPPPQ-----PMPEPAP 576
>TC91433
Length = 738
Score = 50.4 bits (119), Expect = 2e-06
Identities = 56/182 (30%), Positives = 76/182 (40%), Gaps = 13/182 (7%)
Frame = +2
Query: 373 LMLASNCHIDQAIPSDSNPAVPTN-PEHKTPKKWKKRAKKKNVLLSEGGKSNEHNGQPTE 431
L LAS C+ +I + S+ A P+N P +P AK++ V+ + SN+ T
Sbjct: 50 LALASLCYPLASINAQSHAAAPSNLPITTSPFV----AKQQPVVAAISPTSNKPTA--TV 211
Query: 432 TPVQKSVSPINAPSIDPTLPTDQPVPIDLATPKHSEGKMSNAQRNNVLLSEGVKSNEQP- 490
T S P PS TLP + PI ATP + +N Q+ V + +N+QP
Sbjct: 212 TAAAPSKLPTTNPSEPSTLPLYKQQPIVAATPASN----TNKQQPIVAATPASNTNKQPA 379
Query: 491 -----------TETPVQKSVNPIKAPSIDPTLPTDQPVPIDLATPKHSEGKMSKAQRRKK 539
T T S P PS TLP D+ PI ATP S + A
Sbjct: 380 ATSPTSNKPITTVTAAAPSKLPTTNPSAPSTLPLDKQKPIVAATPASSTKQPPAATSPIS 559
Query: 540 NK 541
NK
Sbjct: 560 NK 565
>TC81541 similar to GP|22597168|gb|AAN03471.1 unknown protein {Glycine max},
partial (36%)
Length = 927
Score = 43.5 bits (101), Expect = 2e-04
Identities = 41/176 (23%), Positives = 78/176 (44%), Gaps = 7/176 (3%)
Frame = +3
Query: 33 QSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITLETPKEQGAEPIEATVVH 92
+ C KK R+ K + ++++ ++ VV + I + + G +V
Sbjct: 366 EKCYKKVRKLLNKYPQIRDQNYDDKANIVTITVVCCSPEKIRDKLCYKGGGSIKSIEIVD 545
Query: 93 PVPDQRPAKTHRKKK-KKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAES 151
P P + A+ +KK+ +K EP+ + EPE K+ D +P K+ +K K A+
Sbjct: 546 P-PKPKAAEPEKKKEAEKPKSAEPEKKKEPEKKKEG--DKPKP-----AKEAEKPKAADP 701
Query: 152 NEAVAELATVCTEPPAVQGGVLPEPKPK------EPCSTEHPRVCPEPPLGLAIPI 201
+ V +V + + P+PKP+ +P T P + P+ P +A+P+
Sbjct: 702 EKKVT-FVSVVKDSDKPKDAEKPKPKPEAEKPKDKPAPTAMPMMIPQMPPPMAVPV 866
Score = 29.3 bits (64), Expect = 4.4
Identities = 19/83 (22%), Positives = 36/83 (42%)
Frame = +3
Query: 20 KPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITLETPK 79
+PE E+ + KKK +KKK ++ + E ++ +P T + + ++ K
Sbjct: 567 EPEKKKEAEKPKSAEPEKKKEPEKKKEGDKPKPAKEAEKPKAADPEKKVTFVSVVKDSDK 746
Query: 80 EQGAEPIEATVVHPVPDQRPAKT 102
+ AE + P +PA T
Sbjct: 747 PKDAEKPKPKPEAEKPKDKPAPT 815
>TC91597 similar to GP|21323350|dbj|BAB97978. Hypothetical protein
{Corynebacterium glutamicum ATCC 13032}, partial (5%)
Length = 718
Score = 42.0 bits (97), Expect = 7e-04
Identities = 41/185 (22%), Positives = 76/185 (40%), Gaps = 13/185 (7%)
Frame = +2
Query: 397 PEHKTPKKWKKRAKKKNV-LLSEGGKSNEHNGQPTETPVQKSVSPINAPSIDPTLPTDQP 455
P K+P K + K N L ++ S + G+ + + +P P T T +
Sbjct: 74 PRSKSPSKGQPSRKSSNPQLATQASDSKDTKGRKVDVVAE---APELGPPAALTKETKES 244
Query: 456 VPIDLATPKHS-EGKMSNAQRNNVLL------SEGVKSNEQPTETPVQKSVNPIKAPS-- 506
V P E K Q + V+ +E+P E P++K PIK P+
Sbjct: 245 VEKQEEKPATKLEEKPVEKQEKPIKKPAKKQEDRPVEKHEKPVEEPIEKQEKPIKKPAKK 424
Query: 507 -IDPTLPTDQPVPIDLATPKHSEGKMSKAQRR--KKNKRALKSAWSESVHHSSDQNAKTP 563
D + + P++ K E ++K++++ +K ++ +K + H S ++ K P
Sbjct: 425 QEDRPIEKQEKKPVEKHEKKPVEEPIAKSEKKSAEKLEKPIKKPVEKQDHRSVEKQEKNP 604
Query: 564 VQRTE 568
V+ E
Sbjct: 605 VEEHE 619
Score = 35.4 bits (80), Expect = 0.062
Identities = 29/114 (25%), Positives = 52/114 (45%), Gaps = 4/114 (3%)
Frame = +2
Query: 17 REEKP--EPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPIT 74
+ EKP EP+ E+ ++ K++ R +K +++ +E HE V EP+ +
Sbjct: 356 KHEKPVEEPIEKQEKPIKKPAKKQEDRPIEKQEKKPVEKHEKKPV--EEPIAKS------ 511
Query: 75 LETPKEQGAEPIEATVVHPV--PDQRPAKTHRKKKKKETVLEPQPEPEPEPKET 126
+++ AE +E + PV D R + K +E P + E +P ET
Sbjct: 512 ----EKKSAEKLEKPIKKPVEKQDHRSVEKQEKNPVEEHEKTPVEKHEKKPAET 661
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 40.4 bits (93), Expect = 0.002
Identities = 43/160 (26%), Positives = 56/160 (34%), Gaps = 2/160 (1%)
Frame = +1
Query: 72 PITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDH 131
P PKE+ + +P P++ K R +KK+ QP P P P
Sbjct: 22 PAPKRPPKEKDTQ-----ARNPQPEEATKKQKRAPEKKKNPPRGQPPPPPPPPP-----Q 171
Query: 132 SEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCP 191
+ PT K R+ ++N T PP P PK P + PR P
Sbjct: 172 APPT------KPPTRQTPKNNTPPPPPHT----PPDPTPPQPPPQPPKPPHHEKRPRT-P 318
Query: 192 EPPLGLAIPIDPEKNRKEAEPNESNAE--HPRACPEPPLG 229
EPP G P K + A PR P PP G
Sbjct: 319 EPPGGRPPPPTTPKQARPTTQERGGARGARPRPPPAPPQG 438
Score = 34.3 bits (77), Expect = 0.14
Identities = 34/121 (28%), Positives = 47/121 (38%), Gaps = 5/121 (4%)
Frame = +1
Query: 14 KRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPI 73
KR +EK +P Q +E + +K+ +KKK G P P P
Sbjct: 31 KRPPKEKDTQARNP-QPEEATKKQKRAPEKKKNPPRGQPPPPP-----PPPPQAPPTKPP 192
Query: 74 TLETPKEQGAEPIEATVVHPVPDQ---RPAKTHRKKKKKETVLEP--QPEPEPEPKETSN 128
T +TPK P T P P Q +P K +K+ T P +P P PK+
Sbjct: 193 TRQTPKNNTPPPPPHTPPDPTPPQPPPQPPKPPHHEKRPRTPEPPGGRPPPPTTPKQARP 372
Query: 129 T 129
T
Sbjct: 373 T 375
Score = 31.2 bits (69), Expect = 1.2
Identities = 30/138 (21%), Positives = 38/138 (26%), Gaps = 4/138 (2%)
Frame = +3
Query: 115 PQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLP 174
P P P+ P+ + PT K KK + E P
Sbjct: 15 PTPSPQATPQRKGHPSQKPPTGRSHKKTKKSPRKKEK----------------------P 128
Query: 175 EPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRACPEPPLGLA--- 231
P+P P P P PP +P+K P H P P
Sbjct: 129 PPRPATPPPAPAP---PGPPNQTTNTTNPKKQHTPTTPTHPPRPHTPPTPPPTTQTTPPR 299
Query: 232 -IPIDPEKNKGENLTPTD 248
P DP +G T D
Sbjct: 300 EAPTDPGTTRGTPPTADD 353
Score = 30.8 bits (68), Expect = 1.5
Identities = 59/261 (22%), Positives = 74/261 (27%), Gaps = 27/261 (10%)
Frame = +2
Query: 25 PSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITLETPKEQGAE 84
P PE + K K+ KK K SH + P P P Q +
Sbjct: 56 PKPETPNRKKPQKNKKEPPKKRKTPPEASHPPPRPRPPRP-----------PQPNHQHDK 202
Query: 85 PIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKK 144
P + T HP H T P P P T+ + H ++
Sbjct: 203 PQKTT--HP---------HHPHTPPPTPHPPNPPPNHPNHPTTRSAHGPRNHPGDAPHRR 349
Query: 145 KRKGAESNEAVAELATVCTEPP---AVQGGVLPEPKPKEPCSTEHPRVCPEP-------- 193
+R PP QGG P P P P + E P+P
Sbjct: 350 RRPNKPG------------PPPRRGGGQGGRAPAPPPPPPKAREGRTPKPDPGSKENHNR 493
Query: 194 ---PLGLAIPIDPEKNRKEAEPNESNAEHPRA---CPEPP--------LGLAIPIDPEKN 239
G A PEK P RA P PP G P P+KN
Sbjct: 494 TPGKEGPATHGPPEKKTPPPTPKRRGKHTKRAPGGGPPPPHPGRGGGGPGGPPPPPPKKN 673
Query: 240 --KGENLTPTDENELDRRNRR 258
+G P R+N R
Sbjct: 674 SERGGRAAPRGRPGTKRKNPR 736
>BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophila
melanogaster}, partial (2%)
Length = 734
Score = 40.4 bits (93), Expect = 0.002
Identities = 39/192 (20%), Positives = 65/192 (33%), Gaps = 8/192 (4%)
Frame = +1
Query: 76 ETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVL-----EPQPEPEPEPKETSNTD 130
ETP + E T P+ +T +K +KET E + EPE + ++
Sbjct: 157 ETPTKSETNVPETTTTEPIVQTDSEETPTEKPEKETTTKTTKPEAETTTEPETNTPTKSE 336
Query: 131 HSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPK---PKEPCSTEHP 187
P +++K + G+ + TE + +P+ PK ++E
Sbjct: 337 TDVPETTTTLEKNQNVPGSTTESETETPTKSETETTTTEETPTEKPETDIPKTTTASEKN 516
Query: 188 RVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRACPEPPLGLAIPIDPEKNKGENLTPT 247
+ P +P E+ E E+ P P +PEKNK T
Sbjct: 517 KTTTTEP---TVPTVSEETPTEKPEKETTTTPETEATTQPETDNTPTNPEKNKATTTPET 687
Query: 248 DENELDRRNRRE 259
+ N E
Sbjct: 688 ETTTASETNVSE 723
>TC79702 MtN14
Length = 973
Score = 38.9 bits (89), Expect = 0.006
Identities = 41/172 (23%), Positives = 74/172 (42%), Gaps = 16/172 (9%)
Frame = +2
Query: 2 DSHSQSELLHSKKRRRE--EKPEPLPSPEQDQEQSCSKKK---RRKKKKIKEEGI----- 51
D +Q L +K E E P L + E +E S + K + + K++ EE +
Sbjct: 98 DDTAQLSLKSDEKTTEEIVESPITLAASETKEESSAEEAKPLVKAETKEVVEEVVVPVKD 277
Query: 52 ---ESHEHDQVLLPEPV--VVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKK 106
E+ + +Q EPV + +E KE G +EA P + ++T
Sbjct: 278 TEEETKKEEQTETKEPVEETKENGNSLNVEETKENGDSVVEAVQEKPAEE---SETVNVV 448
Query: 107 KKKETVLEPQPEPEPEPKETSNTDHSEPTQL-CSVKKKKKRKGAESNEAVAE 157
K + V EP+ + + +ETS + E + ++ +KK+ + +N+A E
Sbjct: 449 KDENVVAEPETKDNVKTEETSEEKNEEKVEKEDAMDEKKEEEVITNNDAKIE 604
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 38.5 bits (88), Expect = 0.007
Identities = 38/145 (26%), Positives = 48/145 (32%), Gaps = 3/145 (2%)
Frame = -2
Query: 93 PVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSE--PTQLCSVKKKKKRKGAE 150
P P PA+ R T P P P P ++ P +L ++ +E
Sbjct: 763 PPPQAHPARERRTASLARTPPPPPSSPPPAPLLPPRNGNASLLPRKLVGSPARRSSLSSE 584
Query: 151 SNEAVAELATVCTEPPAVQGGVLPEPK-PKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKE 209
S A LA PP P P+ P EP PR P PP
Sbjct: 583 SLHTPAHLA-----PPRQS----PRPRPPPEPLHNPRPRPPPRPP--------------P 473
Query: 210 AEPNESNAEHPRACPEPPLGLAIPI 234
A P + P A P PP A P+
Sbjct: 472 AAPARAPPSGPPAPPTPPATRAPPV 398
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 38.1 bits (87), Expect = 0.010
Identities = 39/159 (24%), Positives = 64/159 (39%), Gaps = 1/159 (0%)
Frame = +3
Query: 13 KKRRREEKPEPLPSPEQDQEQSCSKK-KRRKKKKIKEEGIESHEHDQVLLPEPVVVATAI 71
K ++EK + + D ++ KK K +KKK+ KEE ++ E D +
Sbjct: 171 KDDEKKEKKDKEKKDKTDVDEGKDKKDKEKKKKEKKEENVKGEEEDGDEKKDK------- 329
Query: 72 PITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDH 131
+ KE+G E + D K+ + K+KK+ E E E K+ N D
Sbjct: 330 EKKKKEKKEKGKEDKD-------KDGEEKKSKKDKEKKKDKNEDDDEGEDGSKKKKNKD- 485
Query: 132 SEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQG 170
KK+KK++ E E + + E A +G
Sbjct: 486 ---------KKEKKKEEDEKEEGKVSVRDIDIEETAKEG 575
Score = 33.5 bits (75), Expect = 0.23
Identities = 28/121 (23%), Positives = 52/121 (42%)
Frame = +3
Query: 13 KKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIP 72
+K+++E+K + ++D E+ SKK + KKK E+ E + + +
Sbjct: 330 EKKKKEKKEKGKEDKDKDGEEKKSKKDKEKKKDKNEDDDEGEDGSKKKKNKD-------- 485
Query: 73 ITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHS 132
E KE+ + V + + AK ++KKKK+ E + E K+ S D
Sbjct: 486 -KKEKKKEEDEKEEGKVSVRDIDIEETAKEGKEKKKKK-------EDKEEKKKKSGKDKD 641
Query: 133 E 133
+
Sbjct: 642 Q 644
>BQ144301 similar to GP|15277263|db alternative name: G2~unknown function
{Homo sapiens}, partial (1%)
Length = 1250
Score = 37.0 bits (84), Expect = 0.021
Identities = 27/132 (20%), Positives = 52/132 (38%)
Frame = -3
Query: 13 KKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIP 72
KK +++ K P +P+ + + S ++++ I G H++ P P+ ++T P
Sbjct: 633 KKNQQKNKITPTKTPQSNNKPKTSAPNKKRRSPIS--G*IRHQYISTRHPHPIFLSTPAP 460
Query: 73 ITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHS 132
P+ + P P P + K + PQP P P +T +
Sbjct: 459 -----------PPLSSASP*PQPPSPPQSLYPSPSK----MNPQPTPNPSSAKTQLYESM 325
Query: 133 EPTQLCSVKKKK 144
P + S K ++
Sbjct: 324 VPNEKQSNKHRQ 289
Score = 32.7 bits (73), Expect = 0.40
Identities = 45/175 (25%), Positives = 71/175 (39%), Gaps = 7/175 (4%)
Frame = -3
Query: 366 NAKLPETLMLASN--CHIDQAIPSDSNPAVPTN----PEHKTPKKWKKRAKKKNVLLSEG 419
N P +A N H D +I S + P P P+ +K+ ++KN +
Sbjct: 774 NTSAPNNAHVAHNNKSHRDLSIYSHNVPQHNNTRV*KPTRAQPRNNRKKNQQKNKITPT- 598
Query: 420 GKSNEHNGQP-TETPVQKSVSPINAPSIDPTLPTDQPVPIDLATPKHSEGKMSNAQRNNV 478
K+ + N +P T P +K SPI+ + T P PI L+TP + +S+A
Sbjct: 597 -KTPQSNNKPKTSAPNKKRRSPISG*IRHQYISTRHPHPIFLSTP--APPPLSSASP*PQ 427
Query: 479 LLSEGVKSNEQPTETPVQKSVNPIKAPSIDPTLPTDQPVPIDLATPKHSEGKMSK 533
S P++ Q + NP A T + VP + + KH + M K
Sbjct: 426 PPSPPQSLYPSPSKMNPQPTPNPSSA----KTQLYESMVPNEKQSNKHRQ*NMEK 274
>TC82158 weakly similar to GP|19920013|gb|AAM08453.1 hypothetical protein
{Dictyostelium discoideum}, partial (4%)
Length = 1230
Score = 36.6 bits (83), Expect = 0.028
Identities = 34/132 (25%), Positives = 48/132 (35%)
Frame = +2
Query: 432 TPVQKSVSPINAPSIDPTLPTDQPVPIDLATPKHSEGKMSNAQRNNVLLSEGVKSNEQPT 491
T KS+S I + P PT P ATP HS ++ S + ++ P
Sbjct: 5 TSTSKSISTIPTSTF-PATPT-HPTTTSPATPAHSTSSTASTPSETPTHSRPITAST*PA 178
Query: 492 ETPVQKSVNPIKAPSIDPTLPTDQPVPIDLATPKHSEGKMSKAQRRKKNKRALKSAWSES 551
TP + P PT T LATP HS S + ++ S S
Sbjct: 179 -TPTHSTSTTTSTPHATPTNSTSTTTSNPLATPTHSTHVTS---------TSTSTSISNS 328
Query: 552 VHHSSDQNAKTP 563
+H S+ + P
Sbjct: 329 TYHVSNHSYPRP 364
Score = 31.6 bits (70), Expect = 0.89
Identities = 25/99 (25%), Positives = 39/99 (39%), Gaps = 5/99 (5%)
Frame = +2
Query: 385 IPSDSNPAVPTNPEHKTPKKWKKRAKKKNVLLSEGGKSNEHNGQPTE-----TPVQKSVS 439
IP+ + PA PT+P +P A + S ++ H+ T TP + +
Sbjct: 32 IPTSTFPATPTHPTTTSPAT---PAHSTSSTASTPSETPTHSRPITAST*PATPTHSTST 202
Query: 440 PINAPSIDPTLPTDQPVPIDLATPKHSEGKMSNAQRNNV 478
+ P PT T LATP HS S + ++
Sbjct: 203 TTSTPHATPTNSTSTTTSNPLATPTHSTHVTSTSTSTSI 319
>AW685995 similar to PIR|I51618|I516 nucleolar phosphoprotein - African
clawed frog, partial (5%)
Length = 516
Score = 36.6 bits (83), Expect = 0.028
Identities = 27/115 (23%), Positives = 49/115 (42%), Gaps = 9/115 (7%)
Frame = +3
Query: 79 KEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEP------EPEPKETSNTDHS 132
K + EP + V ++ AK +KKK++ E +P P +E+S+++
Sbjct: 45 KVKAEEPKKVEKVEKTKGKKAAKAEEEKKKQQKKKEEKPAPMEVEEDSSSSEESSSSEDE 224
Query: 133 EPTQLCSVKKK---KKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCST 184
P + + KK KK +E + +E + E PA + E +E S+
Sbjct: 225 APAKKAAPAKKAAAKKESSSEEESSSSESESEEEEKPAKKAAAKKESSSEEESSS 389
Score = 32.0 bits (71), Expect = 0.68
Identities = 27/141 (19%), Positives = 56/141 (39%)
Frame = +3
Query: 12 SKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAI 71
+ K+ + E+P+ + E+ + + +K + KKK+ K++ + P P+ V
Sbjct: 36 TSKKVKAEEPKKVEKVEKTKGKKAAKAEEEKKKQQKKKEEK---------PAPMEVEEDS 188
Query: 72 PITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDH 131
+ E+ + D+ PAK KK E E E E+ + +
Sbjct: 189 SSSEESSSSE--------------DEAPAKKAAPAKKAAAKKESSSEEESSSSESESEEE 326
Query: 132 SEPTQLCSVKKKKKRKGAESN 152
+P + + KK+ + S+
Sbjct: 327 EKPAKKAAAKKESSSEEESSS 389
>TC83149 similar to GP|13561982|gb|AAK30594.1 flagelliform silk protein
{Argiope trifasciata}, partial (3%)
Length = 1177
Score = 36.6 bits (83), Expect = 0.028
Identities = 39/185 (21%), Positives = 62/185 (33%), Gaps = 2/185 (1%)
Frame = +3
Query: 99 PAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAEL 158
P+ + ++ + +T L P P P P+ H +P Q S + ++ K + S+ A L
Sbjct: 123 PSTSPSRRPRSKTPLPPPPPPLPQ--------HPKPPQK-SPPRMRRSKPSPSSPAATPL 275
Query: 159 ATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIP--IDPEKNRKEAEPNESN 216
T PP P P ST P PP +P + P P
Sbjct: 276 WPQRTTPPPSPSTPRPSP*TPATPSTSPTAPLPTPPPRTTLPPALTPRPPSPSTPPTPKP 455
Query: 217 AEHPRACPEPPLGLAIPIDPEKNKGENLTPTDENELDRRNRREELVALVTSDEKIVNPRD 276
+ P +P P P R RR V+ + ++ +P
Sbjct: 456 GPVSVSPASPSATQRVPWRPTARASSTKVPAAAKP*RRATRRPSAVSRSSRVRRLTHPSL 635
Query: 277 HLVPL 281
VP+
Sbjct: 636 LPVPV 650
>TC77435 similar to GP|7211427|gb|AAF40306.1| RNA helicase {Vigna radiata},
partial (67%)
Length = 1828
Score = 35.8 bits (81), Expect = 0.047
Identities = 34/146 (23%), Positives = 62/146 (42%), Gaps = 5/146 (3%)
Frame = +1
Query: 36 SKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITLETPKEQGAEP---IEATVVH 92
SK+K+ + I S H + P+ T +++E+PK + ++ ++ VV
Sbjct: 43 SKRKKNPQNPKTHRFIPSRFHSSTMPSLPITNDT---VSMESPKSKSSKKKKLLQNDVVS 213
Query: 93 PVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESN 152
V K KK+K++ + + + S+ E ++ +KKKK+ E
Sbjct: 214 EVEAVSAKKKESSKKRKKS-------SDDDEETKSDISSEEGSRKVKKEKKKKKSKVEDE 372
Query: 153 EAV--AELATVCTEPPAVQGGVLPEP 176
E V E+A +P AV + EP
Sbjct: 373 EIVEEEEVAVKKDDPNAVSNFRISEP 450
Score = 29.6 bits (65), Expect = 3.4
Identities = 23/93 (24%), Positives = 41/93 (43%), Gaps = 8/93 (8%)
Frame = +1
Query: 12 SKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKK---KIKEEGIESHEHDQVLLPEPVVVA 68
SKKR++ + + E+ K K+ KKK K+++E I E V +P V+
Sbjct: 250 SKKRKKSSDDDEETKSDISSEEGSRKVKKEKKKKKSKVEDEEIVEEEEVAVKKDDPNAVS 429
Query: 69 T-----AIPITLETPKEQGAEPIEATVVHPVPD 96
+ + L+ K + PI+A + + D
Sbjct: 430 NFRISEPLRMKLKENKIEALFPIQAMTFNTILD 528
>TC81158 similar to GP|9280664|gb|AAF86533.1| F21B7.15 {Arabidopsis
thaliana}, partial (6%)
Length = 890
Score = 35.8 bits (81), Expect = 0.047
Identities = 27/127 (21%), Positives = 49/127 (38%), Gaps = 4/127 (3%)
Frame = +1
Query: 47 KEEGIESHEHDQVLLPEPVVVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKK 106
K+ + + + P V + T +T+ET G+EP V P+ P +
Sbjct: 22 KQRKQNNSSYPRFAAPIRVSIPTNFTLTMETQNNNGSEPGPNLVTAVTPELMP----QNL 189
Query: 107 KKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELA----TVC 162
V P+P+PE +N +P + + G +E E + T+
Sbjct: 190 VNNNPVAAETPKPQPEKLINNNGSELDPNPFAAETPVAGKLGTSDDEIPIEKSVERITLT 369
Query: 163 TEPPAVQ 169
+PP+V+
Sbjct: 370 GDPPSVE 390
>BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania major}, partial
(1%)
Length = 766
Score = 35.0 bits (79), Expect = 0.081
Identities = 32/153 (20%), Positives = 45/153 (28%), Gaps = 3/153 (1%)
Frame = +1
Query: 78 PKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPE---PEPEPKETSNTDHSEP 134
P P T P A + + T P P P P + ++ P
Sbjct: 217 PSTAAPSPSTCTTTGPTSSSTSASSPAATSPRTTSPSPHSSGTPPAPAPSASRSSTSRPP 396
Query: 135 TQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPP 194
+ + R+ A + A A CT P P P P+ + R P
Sbjct: 397 -------RARARRAASRSSPWATPARACTTAPTSASRQTPPPSPRTAAKPKASRCTPSRS 555
Query: 195 LGLAIPIDPEKNRKEAEPNESNAEHPRACPEPP 227
P +P K A + A PRA P
Sbjct: 556 ---RAPTEPSKMAAAAAAAGTRATRPRATAAAP 645
>BQ150123 similar to GP|21109992|gb| TonB-dependent receptor {Xanthomonas
axonopodis pv. citri str. 306}, partial (2%)
Length = 1131
Score = 35.0 bits (79), Expect = 0.081
Identities = 35/155 (22%), Positives = 62/155 (39%), Gaps = 8/155 (5%)
Frame = +2
Query: 400 KTPKKWKKRAKKKNVLLSEGGKSNEHNGQPTETPVQKSVSPINAPSIDP-------TLPT 452
KT K+ + +K+ GGK + N + + +QK+ A S + T
Sbjct: 35 KTEKRNGREGRKRTEGKGGGGKEGQKNKRERKDTIQKAPRERGAGSGEAKRMEQKSTKKR 214
Query: 453 DQPVPIDLATPKHSEGKMSNAQRNNVLLSEGVKSNEQPTETPVQKSVN-PIKAPSIDPTL 511
+QP P TP + + + QRN+ S ++Q +TP Q N +A + D T
Sbjct: 215 EQPGPRTHGTPPRPDPRSAAPQRNSAEKSRSSSPHQQ--QTPTQGGRNRATRARAKDTTA 388
Query: 512 PTDQPVPIDLATPKHSEGKMSKAQRRKKNKRALKS 546
DQ +++ + +Q +R +S
Sbjct: 389 RADQTYAHVKRATRYTSHEAQSSQEHAAARRDTQS 493
>TC87687 similar to GP|8894548|emb|CAB95829.1 hypothetical protein {Cicer
arietinum}, partial (67%)
Length = 1969
Score = 35.0 bits (79), Expect = 0.081
Identities = 40/167 (23%), Positives = 61/167 (35%), Gaps = 13/167 (7%)
Frame = +3
Query: 116 QPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNE-------------AVAELATVC 162
QP+P PEP E +NT +E S K G+ E A+AEL +
Sbjct: 117 QPKPNPEPVE-NNTTETEEVSKPSGDDKIPESGSFKEESTIVGELPEAEKKALAELKQLI 293
Query: 163 TEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRA 222
E P P + P PE P E N+K+ + +E+ +
Sbjct: 294 QEALNKHEFSAPASTTPSPVKEQKPEPTPEAPA-------EETNKKDEQVSETESVVAVT 452
Query: 223 CPEPPLGLAIPIDPEKNKGENLTPTDENELDRRNRREELVALVTSDE 269
+ + P P + + E P +E E ++E VA + DE
Sbjct: 453 TADDDVSTTPPPPPTEAEAEAEQPKEEVE-----KKETEVAASSVDE 578
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 34.7 bits (78), Expect = 0.11
Identities = 78/418 (18%), Positives = 154/418 (36%), Gaps = 38/418 (9%)
Frame = +3
Query: 60 LLPEPVVVATAIPITLETPKEQGAEPIEAT--VVHPVPDQRPAKTHRKKKKKETVLEPQP 117
L P P + +++ + ++ A ++A VV PV + K +++ K + Q
Sbjct: 57 LPPHPPISHSSLLFIMPKSSKKSATKVDAAPVVVSPVKSGKKGKRQAEEEVKAVSAKKQK 236
Query: 118 EPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATV-----CTEPPAVQGGV 172
E K+ + V KK++ ES+E+ E V + PA +G V
Sbjct: 237 VEEVAAKQKA----------LKVVKKEESSSEESSESEDEQPVVKAPAPSKKTPAKKGNV 386
Query: 173 -------LPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKN-----RKEAEPNESNAEHP 220
E + S++ V + P+ A+P KN +K E ++E
Sbjct: 387 KKAQPETTSEESDSDSSSSDEEEV--KKPVSKAVP---SKNGSAPAKKVDTSEEEDSEES 551
Query: 221 RACPEPPLGLAIPID----PEKNKGENLTPTDENELDRRNRREELVALVTSDEKIVNPRD 276
+ P A+P P K + TDE+ D E+ A K V ++
Sbjct: 552 SDEDKKPAAKAVPSKNGSAPAKKAASDEEDTDESS-DEDEEDEKPAA------KAVPSKN 710
Query: 277 HLVPLKSIPGFVLEQQTQPG------PVIPVCPNSASLIGQSQTIPVVDTEQK------- 323
VP K ++ ++ P N ++ ++ + +++++
Sbjct: 711 GSVPAKKADTESSDEDSESSDEEDKKPAAKASKNVSAPTKKAASSSDEESDEESDEDEDA 890
Query: 324 --INKRSRMSKMKRKRMNSMPDKHNTEPPETKKRVLDREEPNKCNAKLPETLMLASNCHI 381
++K + ++K +K + D+ + + K+ + ++ +K N
Sbjct: 891 KPVSKPAAVAKKSKKDSSDSDDEDDDSSSDEDKKPVAAKKEDKMNVD------------- 1031
Query: 382 DQAIPSDSNPAVPTNPEHKTPKKWKKRAKKKNVLLSEGGKSNEHNGQPTETPVQKSVS 439
SD + + KTP+K K K+V + + GKS + TP++ S S
Sbjct: 1032KDGSDSDQSEEESEDEPSKTPQK-----KIKDVEMVDAGKSGKKAPNTPATPIENSGS 1190
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.306 0.126 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,917,312
Number of Sequences: 36976
Number of extensions: 294721
Number of successful extensions: 2518
Number of sequences better than 10.0: 153
Number of HSP's better than 10.0 without gapping: 1892
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2360
length of query: 586
length of database: 9,014,727
effective HSP length: 101
effective length of query: 485
effective length of database: 5,280,151
effective search space: 2560873235
effective search space used: 2560873235
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0148.5


