

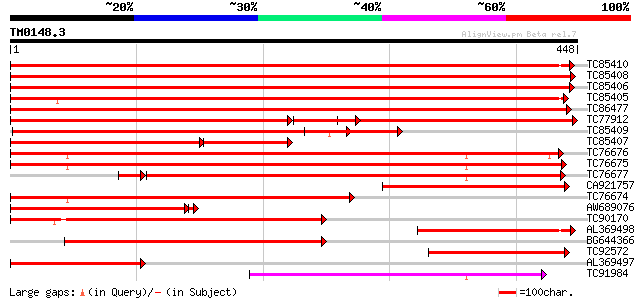
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0148.3
(448 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85410 homologue to PIR|JC2510|JC2510 beta-tubulin R1623 - rice... 878 0.0
TC85408 homologue to PIR|S20869|S20869 tubulin beta-2 chain - ga... 876 0.0
TC85406 SP|P29500|TBB1_PEA Tubulin beta-1 chain. [Garden pea] {P... 863 0.0
TC85405 SP|Q39445|TBB_CICAR Tubulin beta chain. [Chickpea Garba... 845 0.0
TC86477 homologue to GP|15408819|dbj|BAB64211. tubulin beta-4 ch... 837 0.0
TC77912 homologue to GP|10176853|dbj|BAB10059. beta tubulin {Ara... 460 0.0
TC85409 homologue to PIR|S20869|S20869 tubulin beta-2 chain - ga... 472 e-133
TC85407 SP|P28551|TBB3_SOYBN Tubulin beta chain (Fragment). [Soy... 303 e-118
TC76676 homologue to SP|P20363|TBA3_ARATH Tubulin alpha-3/alpha-... 390 e-109
TC76675 SP|P46259|TBA1_PEA Tubulin alpha-1 chain. [Garden pea] {... 384 e-107
TC76677 homologue to SP|P33629|TBA_PRUDU Tubulin alpha chain. [A... 288 1e-80
CA921757 homologue to SP|Q41784|TBB7 Tubulin beta-7 chain (Beta-... 286 1e-77
TC76674 homologue to GP|23452339|gb|AAN33000.1 alpha-tubulin 4 {... 269 2e-72
AW689076 homologue to PIR|JA0048|JA0 tubulin beta-1 chain - soyb... 266 3e-71
TC90170 homologue to GP|12963226|gb|AAK11179.1 alpha-tubulin {Co... 241 4e-64
AL369498 homologue to GP|4079637|emb beta-tubulin {Tetrahymena p... 218 4e-57
BG644366 GP|17402471|em alpha-tubulin {Nicotiana tabacum}, parti... 214 7e-56
TC92572 similar to SP|Q9ZRB0|TBB3_WHEAT Tubulin beta-3 chain (Be... 201 5e-52
AL369497 homologue to SP|O04386|TBB_ Tubulin beta chain. {Chlamy... 201 6e-52
TC91984 homologue to GP|16611919|gb|AAL27406.1 alpha-tubulin {Ar... 188 3e-48
>TC85410 homologue to PIR|JC2510|JC2510 beta-tubulin R1623 - rice, complete
Length = 2139
Score = 878 bits (2269), Expect = 0.0
Identities = 424/446 (95%), Positives = 438/446 (98%)
Frame = +2
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGNTDMQLERVNVYYNEASCGRFV 60
MREILH+QGGQCGNQIG+KFWEVVC EHGID TGRY GN+D+QLERVNVYYNEASCGRFV
Sbjct: 95 MREILHVQGGQCGNQIGSKFWEVVCDEHGIDPTGRYVGNSDLQLERVNVYYNEASCGRFV 274
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
PRAVLMDLEPGTMDSVR+GPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV
Sbjct: 275 PRAVLMDLEPGTMDSVRTGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 454
Query: 121 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV
Sbjct: 455 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 634
Query: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL
Sbjct: 635 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 814
Query: 241 RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDAKNMM 300
RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWD+KNMM
Sbjct: 815 RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDSKNMM 994
Query: 301 CAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
CAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKS+VCDI P G
Sbjct: 995 CAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSSVCDIAPRG 1174
Query: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
L MASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS
Sbjct: 1175LSMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 1354
Query: 421 EYQQYQDATADDEGYDYEDEDEVQEE 446
EYQQYQDATAD+EG +Y+DE+E +++
Sbjct: 1355EYQQYQDATADEEG-EYDDEEEEEQD 1429
>TC85408 homologue to PIR|S20869|S20869 tubulin beta-2 chain - garden pea
(fragment), complete
Length = 2121
Score = 876 bits (2263), Expect = 0.0
Identities = 423/447 (94%), Positives = 438/447 (97%)
Frame = +2
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGNTDMQLERVNVYYNEASCGRFV 60
MREILHIQGGQCGNQIGAKFWEVVCAEHGID TGRY G++D+QLER+NVYYNEAS GRFV
Sbjct: 431 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQLERINVYYNEASGGRFV 610
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV
Sbjct: 611 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 790
Query: 121 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
RKEAEN DCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV
Sbjct: 791 RKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 970
Query: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL
Sbjct: 971 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 1150
Query: 241 RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDAKNMM 300
RFPGQLNSDLRKLAVNL+PFPRLHFFM+GFAPLTSRGSQQYRAL+VPE+TQQMWD+KNMM
Sbjct: 1151RFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKNMM 1330
Query: 301 CAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
CAADPRHGRYLTASA+FRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG
Sbjct: 1331CAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG 1510
Query: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS
Sbjct: 1511LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 1690
Query: 421 EYQQYQDATADDEGYDYEDEDEVQEED 447
EYQQYQDATA+++ Y+ E+ED QE D
Sbjct: 1691EYQQYQDATAEEDEYEDEEEDYQQEHD 1771
>TC85406 SP|P29500|TBB1_PEA Tubulin beta-1 chain. [Garden pea] {Pisum
sativum}, partial (98%)
Length = 1745
Score = 863 bits (2230), Expect = 0.0
Identities = 417/446 (93%), Positives = 433/446 (96%)
Frame = +2
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGNTDMQLERVNVYYNEASCGRFV 60
MR+ILHIQGGQCGNQIGAKFWEVVCAEHGID TGRY G++D+QLER++VYYNEAS GRFV
Sbjct: 116 MRQILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYTGDSDLQLERIDVYYNEASGGRFV 295
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
PRAVLMDLEPGTMDS+RSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV
Sbjct: 296 PRAVLMDLEPGTMDSIRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 475
Query: 121 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV
Sbjct: 476 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 655
Query: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
EPYNATLSVHQLVENADE MVLDNEALYDICFR LKL+ PSFGDLNHLISATMSGVTCCL
Sbjct: 656 EPYNATLSVHQLVENADEVMVLDNEALYDICFRILKLSNPSFGDLNHLISATMSGVTCCL 835
Query: 241 RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDAKNMM 300
RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYR L+VPELTQQMWDAKNMM
Sbjct: 836 RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRTLSVPELTQQMWDAKNMM 1015
Query: 301 CAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
CAADPRHGRYLTASAMFRGKMSTKEVDEQM+NVQNKNSSYFVEWIPNNVKSTVCDIPPTG
Sbjct: 1016CAADPRHGRYLTASAMFRGKMSTKEVDEQMMNVQNKNSSYFVEWIPNNVKSTVCDIPPTG 1195
Query: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLV+
Sbjct: 1196LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVA 1375
Query: 421 EYQQYQDATADDEGYDYEDEDEVQEE 446
EYQQYQDATAD++ Y E+ +E E+
Sbjct: 1376EYQQYQDATADEDEYGEEEGEEDYEQ 1453
>TC85405 SP|Q39445|TBB_CICAR Tubulin beta chain. [Chickpea Garbanzo] {Cicer
arietinum}, partial (98%)
Length = 1840
Score = 845 bits (2182), Expect = 0.0
Identities = 408/443 (92%), Positives = 429/443 (96%), Gaps = 2/443 (0%)
Frame = +2
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRY--QGNTDMQLERVNVYYNEASCGR 58
MREILH+QGGQCGNQIG+KFWEV+C EHGID TG+Y +G D QLER+NVYYNEAS GR
Sbjct: 230 MREILHVQGGQCGNQIGSKFWEVICDEHGIDPTGKYVSEGGNDTQLERINVYYNEASGGR 409
Query: 59 FVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
+VPRAVLMDLEPGTM+S+RSGP+G+IFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD
Sbjct: 410 YVPRAVLMDLEPGTMESIRSGPFGKIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 589
Query: 119 VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT
Sbjct: 590 VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 769
Query: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKL+TPSFGDLNHLISATMSGVTC
Sbjct: 770 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLSTPSFGDLNHLISATMSGVTC 949
Query: 239 CLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDAKN 298
CLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQY +LTVPELTQQMWDAKN
Sbjct: 950 CLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYVSLTVPELTQQMWDAKN 1129
Query: 299 MMCAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPP 358
MMCAADPRHGRYLTASAMFRGKMSTKEVDEQ+INVQNKNSSYFVEWIPNNVKS+VCDIPP
Sbjct: 1130MMCAADPRHGRYLTASAMFRGKMSTKEVDEQIINVQNKNSSYFVEWIPNNVKSSVCDIPP 1309
Query: 359 TGLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDL 418
LKM+STFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDL
Sbjct: 1310KNLKMSSTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDL 1489
Query: 419 VSEYQQYQDATADDEGYDYEDED 441
V+EYQQYQDA A++E DYE+E+
Sbjct: 1490VAEYQQYQDAIAEEED-DYEEEE 1555
>TC86477 homologue to GP|15408819|dbj|BAB64211. tubulin beta-4 chain {Oryza
sativa (japonica cultivar-group)}, partial (98%)
Length = 1816
Score = 837 bits (2163), Expect = 0.0
Identities = 404/445 (90%), Positives = 425/445 (94%), Gaps = 1/445 (0%)
Frame = +1
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGNTDMQ-LERVNVYYNEASCGRF 59
MREILHIQGGQCGNQIG KFWE VC EHGID G+Y GN+ +Q LERVNVYYNE S GR+
Sbjct: 79 MREILHIQGGQCGNQIGTKFWEAVCDEHGIDPIGQYIGNSKLQQLERVNVYYNEGSNGRY 258
Query: 60 VPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDV 119
VPRAVLMDLEPGTMD+VRSGPYGQIFRPDNFVFGQSGAGNN+AKGHYTEGAELID VLDV
Sbjct: 259 VPRAVLMDLEPGTMDAVRSGPYGQIFRPDNFVFGQSGAGNNFAKGHYTEGAELIDFVLDV 438
Query: 120 VRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTV 179
VRKE ENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTV
Sbjct: 439 VRKEVENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTV 618
Query: 180 VEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCC 239
VEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCC
Sbjct: 619 VEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCC 798
Query: 240 LRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDAKNM 299
LRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTS+GSQ YRALTVPELTQQMWDAKNM
Sbjct: 799 LRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSQGSQNYRALTVPELTQQMWDAKNM 978
Query: 300 MCAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPT 359
MCAADPRHGRYLTASA+FRGKMSTKEVDEQ++NVQNKNSSYFVEWIPNNVKS+VCD+PP
Sbjct: 979 MCAADPRHGRYLTASAVFRGKMSTKEVDEQVMNVQNKNSSYFVEWIPNNVKSSVCDVPPR 1158
Query: 360 GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLV 419
GL MASTF+GNSTSIQEMFRRVSEQF+AMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLV
Sbjct: 1159GLSMASTFVGNSTSIQEMFRRVSEQFSAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLV 1338
Query: 420 SEYQQYQDATADDEGYDYEDEDEVQ 444
+EYQQYQDA ++G DY++E++ Q
Sbjct: 1339AEYQQYQDAPVYEDGDDYQEEEDAQ 1413
>TC77912 homologue to GP|10176853|dbj|BAB10059. beta tubulin {Arabidopsis
thaliana}, complete
Length = 1732
Score = 460 bits (1184), Expect(2) = 0.0
Identities = 220/223 (98%), Positives = 221/223 (98%)
Frame = +2
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGNTDMQLERVNVYYNEASCGRFV 60
MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGN D+QLERVNVYYNEASCGRFV
Sbjct: 131 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGNNDLQLERVNVYYNEASCGRFV 310
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV
Sbjct: 311 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 490
Query: 121 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV
Sbjct: 491 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 670
Query: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFG 223
EPYNATLSVHQLVENADECM LDNEALYDICFRTLKLTTPSFG
Sbjct: 671 EPYNATLSVHQLVENADECMALDNEALYDICFRTLKLTTPSFG 799
Score = 347 bits (891), Expect(2) = 0.0
Identities = 171/190 (90%), Positives = 177/190 (93%), Gaps = 1/190 (0%)
Frame = +3
Query: 260 FPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDAKNMMCAADPRHGRYLTASAMFRG 319
FP L F G P SQQYRALTVPELTQQMWD+KNMMCAADPRHGRYLTASAMFRG
Sbjct: 909 FPSLAFLHGGICPTHFSWSQQYRALTVPELTQQMWDSKNMMCAADPRHGRYLTASAMFRG 1088
Query: 320 KMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTGLKMASTFIGNSTSIQEMFR 379
KMSTK+VDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTGLKMASTFIGNSTSIQEMFR
Sbjct: 1089 KMSTKKVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTGLKMASTFIGNSTSIQEMFR 1268
Query: 380 RVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATADDEGYDYED 439
RVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATAD+EGYDYED
Sbjct: 1269 RVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATADEEGYDYED 1448
Query: 440 -EDEVQEEDS 448
E+EVQEE++
Sbjct: 1449 EEEEVQEEEA 1478
Score = 106 bits (265), Expect = 2e-23
Identities = 51/53 (96%), Positives = 51/53 (96%)
Frame = +1
Query: 225 LNHLISATMSGVTCCLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRG 277
LNHLISATMSGVTCCLRF GQLNSDLRKLAVNLIPFPR HFFMVGFAPLTSRG
Sbjct: 805 LNHLISATMSGVTCCLRFTGQLNSDLRKLAVNLIPFPRWHFFMVGFAPLTSRG 963
>TC85409 homologue to PIR|S20869|S20869 tubulin beta-2 chain - garden pea
(fragment), partial (68%)
Length = 1099
Score = 472 bits (1214), Expect = e-133
Identities = 233/270 (86%), Positives = 242/270 (89%), Gaps = 2/270 (0%)
Frame = +3
Query: 3 EILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGNTDMQLERVNVYYNEASCGRFVPR 62
EILHIQGGQCGNQIGAKFWEVVCAEHGID TGRY G +++QLER+NVYYNEASCGRFVPR
Sbjct: 174 EILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYSGTSELQLERINVYYNEASCGRFVPR 353
Query: 63 AVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVVRK 122
A+LMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVVRK
Sbjct: 354 AILMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVVRK 533
Query: 123 EAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVVEP 182
EAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVVEP
Sbjct: 534 EAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVVEP 713
Query: 183 YNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCLRF 242
YNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATM V C + F
Sbjct: 714 YNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATM--VWCHMLF 887
Query: 243 PGQLNSDLR--KLAVNLIPFPRLHFFMVGF 270
++ LR + P F VGF
Sbjct: 888 EVPWSAKLRSEEACCESHSIPSSSLFHVGF 977
Score = 161 bits (407), Expect = 5e-40
Identities = 74/77 (96%), Positives = 77/77 (99%)
Frame = +1
Query: 234 SGVTCCLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQM 293
SGVTCCLRFPGQLNSDLRKLAVNLIPFPRLHFFM+GFAPLTSRGSQQYRAL+VPELTQQM
Sbjct: 868 SGVTCCLRFPGQLNSDLRKLAVNLIPFPRLHFFMLGFAPLTSRGSQQYRALSVPELTQQM 1047
Query: 294 WDAKNMMCAADPRHGRY 310
WD+KNMMCAADPRHGRY
Sbjct: 1048WDSKNMMCAADPRHGRY 1098
>TC85407 SP|P28551|TBB3_SOYBN Tubulin beta chain (Fragment). [Soybean]
{Glycine max}, partial (54%)
Length = 766
Score = 303 bits (775), Expect(2) = e-118
Identities = 140/154 (90%), Positives = 150/154 (96%)
Frame = +2
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGNTDMQLERVNVYYNEASCGRFV 60
MREILHIQGGQCGNQIGAKFWEV+C EHGID TG+Y G++++QLER+NVYYNEAS GR+V
Sbjct: 83 MREILHIQGGQCGNQIGAKFWEVICDEHGIDHTGKYNGDSELQLERINVYYNEASGGRYV 262
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
PRAVLMDLEPGTMDSVRSGP+GQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV
Sbjct: 263 PRAVLMDLEPGTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 442
Query: 121 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISK 154
RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLIS+
Sbjct: 443 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISE 544
Score = 142 bits (358), Expect(2) = e-118
Identities = 68/70 (97%), Positives = 69/70 (98%)
Frame = +3
Query: 154 KIREEYPDRMMLTFSVFPSPKVSDTVVEPYNATLSVHQLVENADECMVLDNEALYDICFR 213
KIREEYPDRMMLTFSVFPSPKVSDTVVEPYNATLSVHQLVENADECMVLDNEALYDICFR
Sbjct: 543 KIREEYPDRMMLTFSVFPSPKVSDTVVEPYNATLSVHQLVENADECMVLDNEALYDICFR 722
Query: 214 TLKLTTPSFG 223
TLKL TP+FG
Sbjct: 723 TLKLATPTFG 752
>TC76676 homologue to SP|P20363|TBA3_ARATH Tubulin alpha-3/alpha-5 chain.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (98%)
Length = 1656
Score = 390 bits (1003), Expect = e-109
Identities = 186/449 (41%), Positives = 286/449 (63%), Gaps = 12/449 (2%)
Frame = +3
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGNTDMQL--ERVNVYYNEASCGR 58
MREI+ I GQ G Q+G WE+ C EHGI G ++ + + + N +++E G+
Sbjct: 72 MREIISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDSSVGVAHDAFNTFFSETGSGK 251
Query: 59 FVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
VPRAV +DLEP +D VRSG Y Q+F P+ + G+ A NN+A+GHYT G E++D LD
Sbjct: 252 HVPRAVFVDLEPTVIDEVRSGTYRQLFHPEQLISGKEDAANNFARGHYTVGKEIVDLCLD 431
Query: 119 VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
VRK A+NC LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+++PSP+VS
Sbjct: 432 RVRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 611
Query: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN LIS +S +T
Sbjct: 612 VVEPYNSVLSTHSLLEHTDVAVLLDNEAIYDICRRSLDIERPTYTNLNRLISQIISSLTT 791
Query: 239 CLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDAKN 298
LRF G +N D+ + NL+P+PR+HF + +AP+ S + L+VPE+T +++ +
Sbjct: 792 SLRFDGAINVDITEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQLSVPEITNAVFEPAS 971
Query: 299 MMCAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPP 358
MM DPRHG+Y+ M+RG + K+V+ + ++ K + FV+W P K + PP
Sbjct: 972 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVGTIKTKRTVQFVDWCPTGFKCGINYQPP 1151
Query: 359 T--------GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
+ ++ A I N+T++ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 1152SVVPGGDLAKVQRAVCMISNNTAVAEVFSRIDHKFDLMYAKRAFVHWYVGEGMEEGEFSE 1331
Query: 411 AESNMNDLVSEYQQY--QDATADDEGYDY 437
A ++ L +Y++ + A D+EG DY
Sbjct: 1332AREDLAALEKDYEEVGAEGAEDDEEGEDY 1418
>TC76675 SP|P46259|TBA1_PEA Tubulin alpha-1 chain. [Garden pea] {Pisum
sativum}, partial (98%)
Length = 1660
Score = 384 bits (987), Expect = e-107
Identities = 182/450 (40%), Positives = 283/450 (62%), Gaps = 10/450 (2%)
Frame = +3
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGNTDMQL--ERVNVYYNEASCGR 58
MRE + I GQ G Q+G WE+ C EHGI G+ G+ + + N +++E G+
Sbjct: 108 MRECISIHIGQAGIQVGNACWELYCLEHGIQPDGQMPGDKTVGGGDDAFNTFFSETGAGK 287
Query: 59 FVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
VPRAV +DLEP +D VR+G Y Q+F P+ + G+ A NN+A+GHYT G E++D LD
Sbjct: 288 HVPRAVFVDLEPTVIDEVRTGAYRQLFHPEQLISGKEDAANNFARGHYTIGKEIVDLCLD 467
Query: 119 VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
+RK A+NC LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+V+PSP+VS +
Sbjct: 468 RIRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTVYPSPQVSTS 647
Query: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN L+S +S +T
Sbjct: 648 VVEPYNSVLSTHSLLEHTDVAVLLDNEAIYDICRRSLDIERPTYTNLNRLVSQVISSLTA 827
Query: 239 CLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDAKN 298
LRF G LN D+ + NL+P+PR+HF + +AP+ S + L+V E+T ++ +
Sbjct: 828 SLRFDGALNVDVTEFQTNLVPYPRIHFMLSSYAPVISAEKAYHEQLSVAEITNSAFEPSS 1007
Query: 299 MMCAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPP 358
MM DPRHG+Y+ M+RG + K+V+ + ++ K + FV+W P K + PP
Sbjct: 1008MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVATIKTKRTIQFVDWCPTGFKCGINYQPP 1187
Query: 359 T--------GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
T ++ A I NSTS+ E+F R+ +F M+ ++AF+HWY GEGM++ EF+E
Sbjct: 1188TVVPGGDLAKVQRAVCMISNSTSVAEVFSRIDHKFDLMYAKRAFVHWYVGEGMEKGEFSE 1367
Query: 411 AESNMNDLVSEYQQYQDATADDEGYDYEDE 440
A ++ L +Y++ + D + D +++
Sbjct: 1368AREDLAALEKDYEEVGAESGDGDDVDGDED 1457
>TC76677 homologue to SP|P33629|TBA_PRUDU Tubulin alpha chain. [Almond
Prunus amygdalus] {Prunus dulcis}, partial (80%)
Length = 1392
Score = 288 bits (738), Expect(2) = 1e-80
Identities = 136/339 (40%), Positives = 214/339 (63%), Gaps = 8/339 (2%)
Frame = +1
Query: 109 GAELIDSVLDVVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFS 168
G E++D LD +RK A+NC LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+
Sbjct: 160 GKEIVDLCLDRIRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFT 339
Query: 169 VFPSPKVSDTVVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHL 228
V+PSP+VS +VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN L
Sbjct: 340 VYPSPQVSTSVVEPYNSVLSTHSLLEHTDVAVLLDNEAIYDICRRSLDIERPTYTNLNRL 519
Query: 229 ISATMSGVTCCLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPE 288
+S +S +T LRF G LN D+ + NL+P+PR+HF + +AP+ S + L+V E
Sbjct: 520 VSQVISSLTASLRFDGALNVDVTEFQTNLVPYPRIHFMLSSYAPVISAEKAYHEQLSVAE 699
Query: 289 LTQQMWDAKNMMCAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNN 348
+T ++ +MM DPRHG+Y+ M+RG + K+V+ + ++ K + FV+W P
Sbjct: 700 ITNSAFEPSSMMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVATIKTKRTIQFVDWCPTG 879
Query: 349 VKSTVCDIPPT--------GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTG 400
K + PPT ++ A I NSTS+ E+F R+ +F M+ ++AF+HWY G
Sbjct: 880 FKCGINYQPPTVVPGGDLAKVQRAVCMISNSTSVAEVFGRIDHKFDLMYAKRAFVHWYVG 1059
Query: 401 EGMDEMEFTEAESNMNDLVSEYQQYQDATADDEGYDYED 439
EGM+E EF+EA ++ L +Y++ + + D +D
Sbjct: 1060EGMEEGEFSEAREDLAALEKDYEEVGAESGEGGDEDMDD 1176
Score = 29.6 bits (65), Expect(2) = 1e-80
Identities = 10/21 (47%), Positives = 15/21 (70%)
Frame = +2
Query: 87 PDNFVFGQSGAGNNWAKGHYT 107
P+ + G+ A NN+A+GHYT
Sbjct: 5 PEQLISGKEDAANNFARGHYT 67
>CA921757 homologue to SP|Q41784|TBB7 Tubulin beta-7 chain (Beta-7 tubulin).
[Maize] {Zea mays}, partial (34%)
Length = 743
Score = 286 bits (731), Expect = 1e-77
Identities = 137/148 (92%), Positives = 144/148 (96%)
Frame = -1
Query: 295 DAKNMMCAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVC 354
D+KNMMCAADPRHG YLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKS+VC
Sbjct: 713 DSKNMMCAADPRHGPYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSSVC 534
Query: 355 DIPPTGLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESN 414
DIPP GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESN
Sbjct: 533 DIPPKGLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESN 354
Query: 415 MNDLVSEYQQYQDATADDEGYDYEDEDE 442
MNDLV+EYQQYQDATAD+E Y+ E+E+E
Sbjct: 353 MNDLVAEYQQYQDATADEEDYEEEEEEE 270
>TC76674 homologue to GP|23452339|gb|AAN33000.1 alpha-tubulin 4 {Gossypium
hirsutum}, partial (90%)
Length = 970
Score = 269 bits (687), Expect = 2e-72
Identities = 125/274 (45%), Positives = 185/274 (66%), Gaps = 2/274 (0%)
Frame = +2
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGNTDMQL--ERVNVYYNEASCGR 58
MRE + + GQ G Q+G WE+ C EHGI G+ + + + N +++E G+
Sbjct: 131 MRECISVHIGQAGIQVGNACWELYCLEHGIGPDGQMPSDKTIGGGDDAFNTFFSETGAGK 310
Query: 59 FVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
VPRAV +DLEP +D VR+G Y Q+F P+ + G+ A NN+A+GHYT G E++D LD
Sbjct: 311 HVPRAVFVDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFARGHYTIGKEIVDLCLD 490
Query: 119 VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
+RK A+NC LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+V+PSP+VS +
Sbjct: 491 RIRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTVYPSPQVSTS 670
Query: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN L+S +S +T
Sbjct: 671 VVEPYNSVLSTHSLLEHTDVAVLLDNEAIYDICRRSLDIERPTYTNLNRLVSQVISSLTA 850
Query: 239 CLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAP 272
LRF G LN D+ + NL+P+PR+HF + +AP
Sbjct: 851 SLRFDGALNVDVTEFQTNLVPYPRIHFMLSSYAP 952
>AW689076 homologue to PIR|JA0048|JA0 tubulin beta-1 chain - soybean, partial
(33%)
Length = 468
Score = 266 bits (679), Expect(2) = 3e-71
Identities = 121/142 (85%), Positives = 132/142 (92%)
Frame = +3
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGNTDMQLERVNVYYNEASCGRFV 60
MREILH+Q GQCGNQIG KFWEV+C EHGID +G Y G + +QLERVNVYYNEAS GR+V
Sbjct: 18 MREILHVQAGQCGNQIGGKFWEVMCDEHGIDPSGSYVGKSHLQLERVNVYYNEASGGRYV 197
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
PRAVLMDLEPGTMDS+RSGP+G+IFRPDNFVFGQ+GAGNNWAKGHYTEGAELIDSVLDVV
Sbjct: 198 PRAVLMDLEPGTMDSLRSGPFGKIFRPDNFVFGQNGAGNNWAKGHYTEGAELIDSVLDVV 377
Query: 121 RKEAENCDCLQGFQVCHSLGGG 142
RKEA NCDCLQGFQ+CHSLGGG
Sbjct: 378 RKEAXNCDCLQGFQICHSLGGG 443
Score = 21.2 bits (43), Expect(2) = 3e-71
Identities = 8/8 (100%), Positives = 8/8 (100%)
Frame = +1
Query: 142 GTGSGMGT 149
GTGSGMGT
Sbjct: 442 GTGSGMGT 465
>TC90170 homologue to GP|12963226|gb|AAK11179.1 alpha-tubulin
{Colletotrichum lagenarium}, partial (56%)
Length = 809
Score = 241 bits (615), Expect = 4e-64
Identities = 119/255 (46%), Positives = 165/255 (64%), Gaps = 5/255 (1%)
Frame = +3
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTG-----RYQGNTDMQLERVNVYYNEAS 55
MRE++ I GQ G QI WE+ C EHGI G R Q + D + +++E
Sbjct: 51 MREVISINVGQAGCQIANSCWELYCLEHGIQPDGYLTEERKQADPD---HGFSTFFSETG 221
Query: 56 CGRFVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDS 115
G++VPRA+ DLEP +D VR+GPY +F P++ + G+ A NN+A+GHYT G ELID
Sbjct: 222 QGKYVPRAIYCDLEPNVVDEVRTGPYRGLFHPEHMITGKEDASNNYARGHYTVGKELIDQ 401
Query: 116 VLDVVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKV 175
VLD VR+ A+NC LQGF V HS GGGTGSG G LL+ ++ +Y + L F V+P+P+
Sbjct: 402 VLDKVRRVADNCVGLQGFLVFHSFGGGTGSGFGALLMERLSVDYGKKSKLEFCVYPAPQT 581
Query: 176 SDTVVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSG 235
+ +VVEPYN+ L+ H +E++D ++DNEA+YDIC R L L P++ +LN LI+ +S
Sbjct: 582 ATSVVEPYNSILTTHTTLEHSDCSFMVDNEAIYDICRRNLGLERPNYENLNRLIAQVVSS 761
Query: 236 VTCCLRFPGQLNSDL 250
+T LRF G LN DL
Sbjct: 762 ITASLRFDGSLNVDL 806
>AL369498 homologue to GP|4079637|emb beta-tubulin {Tetrahymena pyriformis},
partial (27%)
Length = 517
Score = 218 bits (555), Expect = 4e-57
Identities = 104/125 (83%), Positives = 117/125 (93%)
Frame = +3
Query: 323 TKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTGLKMASTFIGNSTSIQEMFRRVS 382
TKEVDEQM+NVQNKNSSYFVEWIPNNVKS+VCDIPP GLKMA TFIGNST+IQE+F+RVS
Sbjct: 3 TKEVDEQMLNVQNKNSSYFVEWIPNNVKSSVCDIPPKGLKMAVTFIGNSTAIQELFKRVS 182
Query: 383 EQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATADDEGYDYEDEDE 442
EQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDL+SEYQQYQDATA+++G ++++E
Sbjct: 183 EQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLISEYQQYQDATAEEDG-EFDEEGG 359
Query: 443 VQEED 447
E D
Sbjct: 360 EGEMD 374
>BG644366 GP|17402471|em alpha-tubulin {Nicotiana tabacum}, partial (47%)
Length = 646
Score = 214 bits (544), Expect = 7e-56
Identities = 99/207 (47%), Positives = 146/207 (69%)
Frame = +1
Query: 44 LERVNVYYNEASCGRFVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAK 103
L N +++E G+ VPRAV +DLEP +D VR+G Y Q+F P+ + G+ A NN+A+
Sbjct: 25 LTAFNTFFSETGAGKHVPRAVFVDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFAR 204
Query: 104 GHYTEGAELIDSVLDVVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRM 163
GHYT G E++D LD +RK A+NC LQGF V +++GGGTGSG+G+LL+ ++ +Y +
Sbjct: 205 GHYTIGKEIVDLCLDRIRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKS 384
Query: 164 MLTFSVFPSPKVSDTVVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFG 223
L F+++PSP+VS +VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++
Sbjct: 385 KLGFTIYPSPQVSTSVVEPYNSVLSTHSLLEHTDVSILLDNEAIYDICRRSLDIERPTYT 564
Query: 224 DLNHLISATMSGVTCCLRFPGQLNSDL 250
+LN LIS +S +T LRF G + D+
Sbjct: 565 NLNRLISQVISSLTASLRFDGAPDVDV 645
>TC92572 similar to SP|Q9ZRB0|TBB3_WHEAT Tubulin beta-3 chain (Beta-3
tubulin). [Wheat] {Triticum aestivum}, partial (25%)
Length = 720
Score = 201 bits (511), Expect = 5e-52
Identities = 95/111 (85%), Positives = 102/111 (91%)
Frame = +1
Query: 332 NVQNKNSSYFVEWIPNNVKSTVCDIPPTGLKMASTFIGNSTSIQEMFRRVSEQFTAMFRR 391
NVQNKNSSYFVEWIPNNVKS+VCDIPPTGL M+STF+GNSTSIQEMFRRVSEQFT MF+R
Sbjct: 1 NVQNKNSSYFVEWIPNNVKSSVCDIPPTGLSMSSTFMGNSTSIQEMFRRVSEQFTVMFKR 180
Query: 392 KAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATADDEGYDYEDEDE 442
KAFLHWYT EGMDEMEFTEAESNMNDLVSEYQQYQDA +EG ED++E
Sbjct: 181 KAFLHWYTAEGMDEMEFTEAESNMNDLVSEYQQYQDAAGVEEGESDEDDEE 333
>AL369497 homologue to SP|O04386|TBB_ Tubulin beta chain. {Chlamydomonas
incerta}, partial (24%)
Length = 414
Score = 201 bits (510), Expect = 6e-52
Identities = 88/107 (82%), Positives = 101/107 (94%)
Frame = +1
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGNTDMQLERVNVYYNEASCGRFV 60
MREI+HIQGGQCGNQIGAKFWEV+ EHG+D TG Y G++D+QLER+NVY+NEA+ GR+V
Sbjct: 94 MREIVHIQGGQCGNQIGAKFWEVISDEHGVDPTGTYHGDSDLQLERINVYFNEATGGRYV 273
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYT 107
PRAVLMDLEPGTMDSVR+GPYGQ+FRPDNFVFGQ+GAGNNWAKGHYT
Sbjct: 274 PRAVLMDLEPGTMDSVRAGPYGQLFRPDNFVFGQTGAGNNWAKGHYT 414
>TC91984 homologue to GP|16611919|gb|AAL27406.1 alpha-tubulin {Artemia
franciscana}, partial (55%)
Length = 755
Score = 188 bits (478), Expect = 3e-48
Identities = 88/243 (36%), Positives = 144/243 (59%), Gaps = 8/243 (3%)
Frame = +1
Query: 190 HQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCLRFPGQLNSD 249
H +E++D ++DNE +YDIC R L + P++ +LN LI+ +S +T LRF G LN D
Sbjct: 4 HTTLEHSDCAFMVDNEGVYDICRRNLDIERPTYTNLNRLIAQVVSSITASLRFDGSLNVD 183
Query: 250 LRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDAKNMMCAADPRHGR 309
L + NL+P+PR+HF +V +AP+ S + LTV E+T ++ N M DPRHG+
Sbjct: 184 LTEFRTNLVPYPRIHFPLVTYAPVISAEKAYHEQLTVAEITYSCFEPNNQMVKCDPRHGK 363
Query: 310 YLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPT--------GL 361
Y+ ++RG + K+V+ + ++ K + FV+W P K + PPT +
Sbjct: 364 YMACCLLYRGDVVPKDVNAAIATIKTKRTIQFVDWCPTGFKVGINYQPPTVVPGGDLAKV 543
Query: 362 KMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSE 421
+ A + N+T+I E + R+ +F M+ ++AF+HWY GEGM+E EF+EA ++ L +
Sbjct: 544 QRAVCMLSNTTAIAEAWARLDHKFDLMYAKRAFVHWYVGEGMEEGEFSEAREDLAALEKD 723
Query: 422 YQQ 424
Y++
Sbjct: 724 YEE 732
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,885,405
Number of Sequences: 36976
Number of extensions: 166154
Number of successful extensions: 985
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 899
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 944
length of query: 448
length of database: 9,014,727
effective HSP length: 99
effective length of query: 349
effective length of database: 5,354,103
effective search space: 1868581947
effective search space used: 1868581947
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0148.3


