

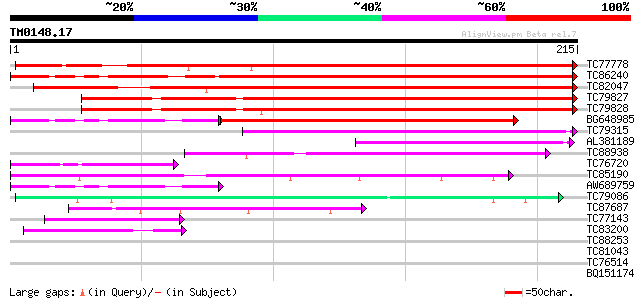
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0148.17
(215 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77778 similar to PIR|T07780|T07780 remorin - potato, partial (... 284 2e-77
TC86240 weakly similar to PIR|T07780|T07780 remorin - potato, pa... 193 5e-50
TC82047 weakly similar to GP|21555669|gb|AAM63910.1 remorin {Ara... 155 1e-38
TC79827 weakly similar to PIR|T46136|T46136 remorin-like protein... 140 4e-34
TC79828 weakly similar to PIR|T07780|T07780 remorin - potato, pa... 135 9e-33
BG648985 weakly similar to PIR|T07780|T07 remorin - potato, part... 129 8e-31
TC79315 similar to PIR|B84847|B84847 hypothetical protein At2g41... 75 2e-14
AL381189 similar to GP|11072019|gb| F12A21.28 {Arabidopsis thali... 64 3e-11
TC88938 similar to PIR|F85436|F85436 hypothetical protein AT4g36... 64 6e-11
TC76720 similar to GP|11121502|emb|CAC14888. putative extensin {... 44 4e-05
TC85190 similar to GP|8894548|emb|CAB95829.1 hypothetical protei... 44 6e-05
AW689759 weakly similar to GP|10727255|gb Shab gene product {alt... 42 1e-04
TC79086 similar to GP|23499033|emb|CAD51113. ubiquitin-protein l... 42 2e-04
TC87687 similar to GP|8894548|emb|CAB95829.1 hypothetical protei... 40 5e-04
TC77143 similar to GP|559557|gb|AAA51649.1|| arabinogalactan-pro... 40 5e-04
TC83200 weakly similar to GP|21554083|gb|AAM63164.1 unknown {Ara... 40 9e-04
TC88253 weakly similar to GP|15810597|gb|AAL07186.1 unknown prot... 39 0.001
TC81043 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproli... 39 0.001
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 38 0.003
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 38 0.003
>TC77778 similar to PIR|T07780|T07780 remorin - potato, partial (68%)
Length = 1103
Score = 284 bits (727), Expect = 2e-77
Identities = 156/216 (72%), Positives = 174/216 (80%), Gaps = 3/216 (1%)
Frame = +3
Query: 3 EEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPP 62
+EQPKK+ESES SNPPPP PA TE+T P EA PK+DVAEEKS+IPQ + P
Sbjct: 267 QEQPKKIESESTSNPPPP-PASTETTTTPLPEA---------PKKDVAEEKSVIPQDNNP 416
Query: 63 ESKP--VDDSKAIVKVEKTQEAAEEKPLEG-SINRDAVLTRVATEKRLSLIKAWEESEKS 119
P VDDSKA+V V+KT EAAEEKP EG SI+RDAVLTRVATEKRLSLIKAWEESEKS
Sbjct: 417 PPPPPVVDDSKALVIVQKTDEAAEEKPKEGGSIDRDAVLTRVATEKRLSLIKAWEESEKS 596
Query: 120 IADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEK 179
A+NKA ++LS I+AWENSK AAKE ELRK+EE LEKKK Y EKLKNKIA +H+ AEEK
Sbjct: 597 KAENKAQRRLSTITAWENSKKAAKEAELRKLEEQLEKKKGEYAEKLKNKIAALHKAAEEK 776
Query: 180 RAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 215
+A IEAKKGEDLLKAEE+AAKYRATGTAPKK F F
Sbjct: 777 KAMIEAKKGEDLLKAEEIAAKYRATGTAPKKLFGLF 884
>TC86240 weakly similar to PIR|T07780|T07780 remorin - potato, partial (65%)
Length = 1140
Score = 193 bits (490), Expect = 5e-50
Identities = 109/215 (50%), Positives = 144/215 (66%)
Frame = +1
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
M E Q K+ +P+ P PAP PT P P A EP + +AP +V E+K++ P P
Sbjct: 157 MAELQTKQEAVPAPA--PAPAPVPT---PVPAAV---EPPLAEAPPVNVVEKKAVAPPPV 312
Query: 61 PPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSI 120
DD+KA+V V+ ++ E S++RD L + EKRLS +KAWE+SEK+
Sbjct: 313 ------ADDTKALVVVDN-EKIPEPVKKNASLDRDIALAEIGKEKRLSNVKAWEDSEKTK 471
Query: 121 ADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKR 180
A+NKA K+LS ++AWENSK AA E +LRKIEE LEKKKA Y EK+KNK+AMVH++AEEKR
Sbjct: 472 AENKAQKQLSTVAAWENSKKAALEAQLRKIEEQLEKKKAEYGEKIKNKVAMVHKQAEEKR 651
Query: 181 AFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 215
A +EA++ E +LKAEE+AAK+ ATGT PKK F
Sbjct: 652 AIVEAQRAEAILKAEEIAAKHNATGTVPKKLLGCF 756
>TC82047 weakly similar to GP|21555669|gb|AAM63910.1 remorin {Arabidopsis
thaliana}, partial (59%)
Length = 827
Score = 155 bits (392), Expect = 1e-38
Identities = 86/209 (41%), Positives = 127/209 (60%), Gaps = 3/209 (1%)
Frame = +1
Query: 10 ESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDD 69
++ESP P P E +P E +++ + SI +P+ +DD
Sbjct: 199 KTESPQLQSLLDPVPEELSPVVEKDSETLSTI------------SISQEPNQQAISTLDD 342
Query: 70 SKAI---VKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAH 126
K ++T + ++K + S +RDA L ++ EKRL+LIKAWEESEK+ A+N+A+
Sbjct: 343 QKVADDHADNKETGDHDDKKDAKDSTDRDAGLAKIVAEKRLALIKAWEESEKTKAENRAY 522
Query: 127 KKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAK 186
KK S + WE SK ++ E +L+K EENLE+KK YV K+KN++A +H+ AEEKRA +EA+
Sbjct: 523 KKQSSVGLWEESKKSSIEAQLKKFEENLERKKVEYVSKMKNELAEIHQYAEEKRAIVEAQ 702
Query: 187 KGEDLLKAEELAAKYRATGTAPKKPFSFF 215
K E+ L+ EE AAK+R+ G APKK F F
Sbjct: 703 KREECLELEETAAKFRSRGVAPKKLFGCF 789
>TC79827 weakly similar to PIR|T46136|T46136 remorin-like protein -
Arabidopsis thaliana, partial (71%)
Length = 776
Score = 140 bits (353), Expect = 4e-34
Identities = 75/188 (39%), Positives = 119/188 (62%)
Frame = +1
Query: 28 TPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIVKVEKTQEAAEEKP 87
TP P++E++P + +++ E + S + + V A V++T A +
Sbjct: 43 TPLPQSESEPREFSYFLEEKEPGNEGT---SSSVVKQERVVSDHATSSVDQTTAAGTDT- 210
Query: 88 LEGSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKEVEL 147
+ S++RDAVL RV ++KRL+LIKAWEE+EK+ +N+A+K S + WE+ K A+ E +
Sbjct: 211 -KDSVDRDAVLARVESQKRLALIKAWEENEKTKVENRAYKMQSAVDLWEDDKKASIEAKF 387
Query: 148 RKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTA 207
+ IE L++KK+ YVE ++NKI +H+ AEEK+A IEA+KGE++LK EE AAK+R G
Sbjct: 388 KGIEVKLDRKKSEYVEVMQNKIGEIHKSAEEKKAMIEAQKGEEILKVEETAAKFRTRGYQ 567
Query: 208 PKKPFSFF 215
P++ F
Sbjct: 568 PRRLLGCF 591
>TC79828 weakly similar to PIR|T07780|T07780 remorin - potato, partial (25%)
Length = 852
Score = 135 bits (341), Expect = 9e-33
Identities = 75/191 (39%), Positives = 119/191 (62%), Gaps = 3/191 (1%)
Frame = +3
Query: 28 TPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIVKVEKTQEAAEEKP 87
TP P++E++P + +++ E + S + + V A V++T A +
Sbjct: 132 TPLPQSESEPREFSYFLEEKEPGNEGT---SSSVVKQERVVSDHATSSVDQTTAAGTDT- 299
Query: 88 LEGSINR---DAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKE 144
+ S++R DAVL RV ++KRL+LIKAWEE+EK+ +N+A+K S + WE+ K A+ E
Sbjct: 300 -KDSVDRVPLDAVLARVESQKRLALIKAWEENEKTKVENRAYKMQSAVDLWEDDKKASIE 476
Query: 145 VELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRAT 204
+ + IE L++KK+ YVE ++NKI +H+ AEEK+A IEA+KGE++LK EE AAK+R
Sbjct: 477 AKFKGIEVKLDRKKSEYVEVMQNKIGEIHKSAEEKKAMIEAQKGEEILKVEETAAKFRTR 656
Query: 205 GTAPKKPFSFF 215
G P++ F
Sbjct: 657 GYQPRRLLGMF 689
>BG648985 weakly similar to PIR|T07780|T07 remorin - potato, partial (58%)
Length = 793
Score = 129 bits (324), Expect = 8e-31
Identities = 64/113 (56%), Positives = 84/113 (73%)
Frame = +2
Query: 81 EAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKI 140
E E S++RD L + EKRLS +KAWE+SEK+ A+NKA K+LS ++AWENSK
Sbjct: 455 EIPEPVKKNASLDRDIALAEIGKEKRLSNVKAWEDSEKTKAENKAQKQLSTVAAWENSKK 634
Query: 141 AAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLK 193
AA E +LRKIEE LEKKKA Y EK+KNK MVH++AEEK++ +EA++ E +LK
Sbjct: 635 AALEAQLRKIEEQLEKKKAEYGEKIKNKGGMVHKQAEEKKSIVEAQRAEAILK 793
Score = 42.4 bits (98), Expect = 1e-04
Identities = 30/81 (37%), Positives = 42/81 (51%)
Frame = +3
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
M E Q K+ +P+ P PAP PT P P A EP + +AP +V E+K++ P P
Sbjct: 60 MAELQTKQEAVPAPA--PAPAPVPT---PVPAAV---EPPLAEAPPVNVVEKKAVAPPP- 212
Query: 61 PPESKPVDDSKAIVKVEKTQE 81
DD+KA+V V+ E
Sbjct: 213 -----VADDTKALVVVDNESE 260
>TC79315 similar to PIR|B84847|B84847 hypothetical protein At2g41870
[imported] - Arabidopsis thaliana, partial (57%)
Length = 1293
Score = 75.1 bits (183), Expect = 2e-14
Identities = 42/127 (33%), Positives = 72/127 (56%)
Frame = +1
Query: 89 EGSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELR 148
+G + RV E+ + I AW+ ++ + +N+ + + I+ WE+ ++ ++
Sbjct: 574 QGGSEEHVSVDRVKKEEVDAKISAWQNAKVAKINNRFKRDDAVINGWESEQVQKATSWMK 753
Query: 149 KIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAP 208
K+E LE+K+A +EK +NKIA R+AEE++A EAK+G + + E+A RA G P
Sbjct: 754 KVERKLEEKRARALEKTQNKIAKARRKAEERKASAEAKRGTKVARVLEIANLMRAVGRPP 933
Query: 209 KKPFSFF 215
K SFF
Sbjct: 934 AKK-SFF 951
>AL381189 similar to GP|11072019|gb| F12A21.28 {Arabidopsis thaliana},
partial (27%)
Length = 520
Score = 64.3 bits (155), Expect = 3e-11
Identities = 36/83 (43%), Positives = 48/83 (57%)
Frame = +1
Query: 132 ISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDL 191
I AWEN +I E+E++K+E E+ KA+ EKL NK+A R AEEKRA + K ++
Sbjct: 22 IQAWENHQIRKAEMEMKKMEVKAERMKAMAQEKLTNKLAATRRIAEEKRANAQVKLNDNA 201
Query: 192 LKAEELAAKYRATGTAPKKPFSF 214
L+ E R TG P FSF
Sbjct: 202 LRTTERVDYIRRTGHVPSS-FSF 267
>TC88938 similar to PIR|F85436|F85436 hypothetical protein AT4g36970
[imported] - Arabidopsis thaliana, partial (28%)
Length = 1701
Score = 63.5 bits (153), Expect = 6e-11
Identities = 39/142 (27%), Positives = 73/142 (50%), Gaps = 3/142 (2%)
Frame = +2
Query: 67 VDDSKAIVKVEKTQEAAEEKPL---EGSINRDAVLTRVATEKRLSLIKAWEESEKSIADN 123
+ D + +V T+ + + K L GS N D+ + + + S WE SE+S +
Sbjct: 860 IRDVQVDERVTMTRWSKKHKALFTGRGSENVDSWKKKETSTRSSS----WEISERSKTVS 1027
Query: 124 KAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFI 183
KA ++ + I+AWEN + A E ++K+E LEKK+A ++K+ NK+ ++A+E R+ +
Sbjct: 1028KAKREEAKITAWENLQKAKAEAAIQKLEMKLEKKRASSMDKIMNKLKFAQKKAQEMRSSV 1207
Query: 184 EAKKGEDLLKAEELAAKYRATG 205
+ + + +R G
Sbjct: 1208SVDQAHQVARTSHKVMSFRRAG 1273
>TC76720 similar to GP|11121502|emb|CAC14888. putative extensin {Nicotiana
sylvestris}, partial (51%)
Length = 1290
Score = 44.3 bits (103), Expect = 4e-05
Identities = 22/64 (34%), Positives = 34/64 (52%)
Frame = +1
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
+ P + + SP+N PP AP PT TP+P +++ P P ++P + S P PS
Sbjct: 592 LNSPSPTPIPTPSPANSPP-APTPTP-TPSPHSDSPPAPSPDNSPSSSPSPSPSSSPAPS 765
Query: 61 PPES 64
P E+
Sbjct: 766 PDEA 777
Score = 30.0 bits (66), Expect = 0.70
Identities = 15/53 (28%), Positives = 23/53 (43%), Gaps = 3/53 (5%)
Frame = +1
Query: 6 PKKVESESPSNPPPPAPAPTES---TPAPEAEAKPEPVVHDAPKEDVAEEKSI 55
P + SP + PPAP+P S +P+P + P P +A + I
Sbjct: 652 PTPTPTPSPHSDSPPAPSPDNSPSSSPSPSPSSSPAPSPDEAADNNAISHTGI 810
Score = 29.6 bits (65), Expect = 0.91
Identities = 20/62 (32%), Positives = 28/62 (44%), Gaps = 1/62 (1%)
Frame = +2
Query: 6 PKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVV-HDAPKEDVAEEKSIIPQPSPPES 64
P K + PS PPP + P + ++ P PV + +P V + KS P PP
Sbjct: 203 PPKKPYKYPSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKS----PPPPPK 370
Query: 65 KP 66
KP
Sbjct: 371 KP 376
Score = 29.6 bits (65), Expect = 0.91
Identities = 20/62 (32%), Positives = 28/62 (44%), Gaps = 1/62 (1%)
Frame = +2
Query: 6 PKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVV-HDAPKEDVAEEKSIIPQPSPPES 64
P K + PS PPP + P + ++ P PV + +P V + KS P PP
Sbjct: 44 PPKKPYKYPSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKS----PPPPPK 211
Query: 65 KP 66
KP
Sbjct: 212 KP 217
>TC85190 similar to GP|8894548|emb|CAB95829.1 hypothetical protein {Cicer
arietinum}, partial (50%)
Length = 1496
Score = 43.5 bits (101), Expect = 6e-05
Identities = 50/206 (24%), Positives = 87/206 (41%), Gaps = 15/206 (7%)
Frame = +3
Query: 1 MTEEQPKKVESESPSNPPPPAPAPT----ESTPAPEAEAKPEPVVHDAPKEDVAEEKSII 56
+ +E K E +P P AP P E E E K E VV + E V EEK +
Sbjct: 468 LIQEALNKHEFTAPPPAPVKAPEPEVAVKEEKKPEEDEKKTEEVVEEKKDEAVVEEKKVD 647
Query: 57 PQPSPPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEK-RLSLIKAWEE 115
+ +P KVE + EEK +E ++ +TE+ ++A +E
Sbjct: 648 EEKGSTSEEP--------KVETAEPEKEEKKVEETVVEVVEKIAASTEEDGAKTVEAIQE 803
Query: 116 SEKSIADNKAHKKLSD-ISAWENSKIAAKEVELRKIEENLEKKKAVYV--------EKLK 166
S S+ + + +++ ++ E + I +EVE+ I +++ V + K+K
Sbjct: 804 SIVSVPVTEGEQPVAEPVAEVEVTPIVPEEVEIWGIPLLADERSDVILLKFLRARDFKVK 983
Query: 167 NKIAMVHREAEEKRAF-IEAKKGEDL 191
M+ + ++ F +EA EDL
Sbjct: 984 EAFTMIKQTVLWRKEFGVEALLQEDL 1061
>AW689759 weakly similar to GP|10727255|gb Shab gene product {alt 1}
[Drosophila melanogaster], partial (2%)
Length = 648
Score = 42.4 bits (98), Expect = 1e-04
Identities = 30/81 (37%), Positives = 42/81 (51%)
Frame = +2
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
M E Q K+ +P+ P PAP PT P P A EP + +AP +V E+K++ P P
Sbjct: 74 MAELQTKQEAVPAPA--PAPAPVPT---PVPAAV---EPPLAEAPPVNVVEKKAVAPPP- 226
Query: 61 PPESKPVDDSKAIVKVEKTQE 81
DD+KA+V V+ E
Sbjct: 227 -----VADDTKALVVVDNESE 274
>TC79086 similar to GP|23499033|emb|CAD51113. ubiquitin-protein ligase 1
putative {Plasmodium falciparum 3D7}, partial (0%)
Length = 1418
Score = 41.6 bits (96), Expect = 2e-04
Identities = 52/239 (21%), Positives = 86/239 (35%), Gaps = 31/239 (12%)
Frame = +2
Query: 3 EEQPKKVESESPSNPPPPAPAP---------TESTPAPEAEAKP-----EPVVHDAPKED 48
EE+ + E E+P PPP P E T E E P E + K
Sbjct: 173 EEEEESSEEEAPKTTPPPKTNPKNNSVEDEDEEETDTDEDEPPPKTKPIEQTLKSGTKPS 352
Query: 49 VAEEKSIIPQPSPPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLS 108
+A +S +P+ + K + + ++ E + + S N+ A RV TE
Sbjct: 353 IAPARSGTKRPAENNDTKQSNKKKTTEEKNKKKEKEPEEDDNSNNKKASFQRVFTEDDEV 532
Query: 109 LIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNK 168
I + + N K LS + K V L ++ + + + + Y K+K+K
Sbjct: 533 AILQGLVDFTAKSGNDPTKHLSAFYQIVKKSVHFK-VTLDQLRDKVRRLRLKYENKIKSK 709
Query: 169 IAMVHREAEEKRAF-----------IEAKKGEDLLKA------EELAAKYRATGTAPKK 210
+ E+ F ++ +GE+ K +ELA K A KK
Sbjct: 710 KTPTFSKPVEETMFELSKKIWGGGKVDVNEGEENEKVNGKPAKKELAVKKAAAARTTKK 886
>TC87687 similar to GP|8894548|emb|CAB95829.1 hypothetical protein {Cicer
arietinum}, partial (67%)
Length = 1969
Score = 40.4 bits (93), Expect = 5e-04
Identities = 32/129 (24%), Positives = 61/129 (46%), Gaps = 16/129 (12%)
Frame = +3
Query: 23 APTESTPAPEAEAKPEPVVHDAPKED-------VAEEKSIIPQPSPPE----SKPVDDSK 71
AP +TP+P E KPEP +AP E+ V+E +S++ + + + P ++
Sbjct: 324 APASTTPSPVKEQKPEP-TPEAPAEETNKKDEQVSETESVVAVTTADDDVSTTPPPPPTE 500
Query: 72 AIVKVEKTQEAAEEKPLE---GSINRDAVLTRVATEKRLSLIKAWEESEKSI--ADNKAH 126
A + E+ +E E+K E S++ D T A E+ + + + E + A +
Sbjct: 501 AEAEAEQPKEEVEKKETEVAASSVDEDGAKTVEAIEESVVAVASSVPEEPKVVEASSPEQ 680
Query: 127 KKLSDISAW 135
++ ++S W
Sbjct: 681 QQPEEVSIW 707
Score = 31.6 bits (70), Expect = 0.24
Identities = 41/206 (19%), Positives = 73/206 (34%), Gaps = 17/206 (8%)
Frame = +3
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTE-STPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQP 59
+T++ V ++ S+ P P P P E +T E +KP EE +I+ +
Sbjct: 69 VTQKTESPVPADKESDQPKPNPEPVENNTTETEEVSKPSGDDKIPESGSFKEESTIVGEL 248
Query: 60 SPPESKPVDDSKAIVK----VEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEE 115
E K + + K +++ + A P + E+ + E
Sbjct: 249 PEAEKKALAELKQLIQEALNKHEFSAPASTTPSPVKEQKPEPTPEAPAEETNKKDEQVSE 428
Query: 116 SEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKK------------AVYVE 163
+E +A A D+S E E + +E +EKK+ A VE
Sbjct: 429 TESVVAVTTAD---DDVSTTPPPPPTEAEAEAEQPKEEVEKKETEVAASSVDEDGAKTVE 599
Query: 164 KLKNKIAMVHREAEEKRAFIEAKKGE 189
++ + V E+ +EA E
Sbjct: 600 AIEESVVAVASSVPEEPKVVEASSPE 677
>TC77143 similar to GP|559557|gb|AAA51649.1|| arabinogalactan-protein {Pyrus
communis}, partial (30%)
Length = 956
Score = 40.4 bits (93), Expect = 5e-04
Identities = 20/53 (37%), Positives = 22/53 (40%)
Frame = +3
Query: 14 PSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKP 66
P+ PPPA APT +TPAP DAP D P P P P
Sbjct: 282 PTATPPPAAAPTPATPAPATSPPAPTPTSDAPTPDSTSSSPPAPGPGGPAPGP 440
Score = 32.0 bits (71), Expect = 0.18
Identities = 21/67 (31%), Positives = 26/67 (38%), Gaps = 2/67 (2%)
Frame = +3
Query: 6 PKKVESESPSNPPPPAPAPTESTPAPEAEAKPE--PVVHDAPKEDVAEEKSIIPQPSPPE 63
P + S +P+ PP PAP T T P P P AP + P P+P
Sbjct: 192 PTQPPSATPT-PPTPAPVATPPTATPPTATPPTATPPPAAAPTPATPAPATSPPAPTPTS 368
Query: 64 SKPVDDS 70
P DS
Sbjct: 369 DAPTPDS 389
Score = 30.4 bits (67), Expect = 0.54
Identities = 19/68 (27%), Positives = 25/68 (35%), Gaps = 1/68 (1%)
Frame = +3
Query: 4 EQPKKVESESPS-NPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPP 62
+ P ++ PS P PP PAP +TP P P + P SPP
Sbjct: 174 QAPGAAPTQPPSATPTPPTPAPV-ATPPTATPPTATPPTATPPPAAAPTPATPAPATSPP 350
Query: 63 ESKPVDDS 70
P D+
Sbjct: 351 APTPTSDA 374
>TC83200 weakly similar to GP|21554083|gb|AAM63164.1 unknown {Arabidopsis
thaliana}, partial (71%)
Length = 565
Score = 39.7 bits (91), Expect = 9e-04
Identities = 18/62 (29%), Positives = 28/62 (45%)
Frame = -3
Query: 6 PKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESK 65
PK+ +++ P PPP +P P P P P P + P + ++ PSP S
Sbjct: 473 PKQPKAQKPXLPPPQSPVPNSQAPPPSLSPSPPPPILVPPSQ-------VLSPPSPILSP 315
Query: 66 PV 67
P+
Sbjct: 314 PI 309
>TC88253 weakly similar to GP|15810597|gb|AAL07186.1 unknown protein
{Arabidopsis thaliana}, partial (51%)
Length = 1122
Score = 39.3 bits (90), Expect = 0.001
Identities = 43/173 (24%), Positives = 77/173 (43%), Gaps = 6/173 (3%)
Frame = +3
Query: 35 AKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIVKVEKTQEAAEEKP------L 88
A P V+ K+ + + I P P P E K ++ K + K +E +E P L
Sbjct: 342 ADPIKVLKRLQKKSGKKVELISPLPKPQEEKKEEEIK---EEPKPEEKKDEPPPVVTIVL 512
Query: 89 EGSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELR 148
+ ++ DA + +KR+ IK E E + +++A K + +K+ + +
Sbjct: 513 KIRMHCDACAQVI--QKRIRKIKGVESVETDLGNDQAIVK----GVIDPTKLVDEVFKRT 674
Query: 149 KIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKY 201
K + ++ KK+ E+ K + + EEK+ E KGED K E ++Y
Sbjct: 675 KKQASIVKKEEKKEEEKKEEEKKEEVKEEEKKESEEENKGEDDNKTEIKRSEY 833
>TC81043 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (19%)
Length = 555
Score = 39.3 bits (90), Expect = 0.001
Identities = 25/76 (32%), Positives = 31/76 (39%), Gaps = 1/76 (1%)
Frame = +1
Query: 5 QPKKVESESPSNPP-PPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPE 63
+P SP +PP PPAP S PAP P P AP + P+PP+
Sbjct: 202 KPPPPAPPSPGSPPKPPAPPSPGSPPAPAPAPPPIPPSPPAPPSPGSPPSPPPASPAPPK 381
Query: 64 SKPVDDSKAIVKVEKT 79
P S + EKT
Sbjct: 382 PTPPKGSPLLPMPEKT 429
Score = 31.6 bits (70), Expect = 0.24
Identities = 14/33 (42%), Positives = 18/33 (54%)
Frame = +1
Query: 13 SPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAP 45
SP +PPP +PAP + TP + P P AP
Sbjct: 340 SPPSPPPASPAPPKPTPPKGSPLLPMPEKTPAP 438
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 38.1 bits (87), Expect = 0.003
Identities = 44/210 (20%), Positives = 73/210 (33%), Gaps = 13/210 (6%)
Frame = +3
Query: 3 EEQPKKVESESPSNPPP-PAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSP 61
EE + SES P APAP++ TPA + K + + D S +
Sbjct: 285 EESSSEESSESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETTSEESDSDSSSSDEEEVKK 464
Query: 62 PESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEK------RLSLIKAWEE 115
P SK V +K + EE E S + K + + +
Sbjct: 465 PVSKAVPSKNGSAPAKKVDTSEEEDSEESSDEDKKPAAKAVPSKNGSAPAKKAASDEEDT 644
Query: 116 SEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLE------KKKAVYVEKLKNKI 169
E S D + K + +N + AK+ + +E+ E KK A K +
Sbjct: 645 DESSDEDEEDEKPAAKAVPSKNGSVPAKKADTESSDEDSESSDEEDKKPAAKASKNVSAP 824
Query: 170 AMVHREAEEKRAFIEAKKGEDLLKAEELAA 199
+ ++ + E+ + ED + AA
Sbjct: 825 TKKAASSSDEESDEESDEDEDAKPVSKPAA 914
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 38.1 bits (87), Expect = 0.003
Identities = 21/68 (30%), Positives = 31/68 (44%), Gaps = 2/68 (2%)
Frame = +2
Query: 14 PSNPPPPAPAPTESTPAPEA--EAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSK 71
P NP PP+ P + TP PE KP+ + PK+ ++ P P P +P +
Sbjct: 11 PPNPQPPSDPPKKRTPKPETPNRKKPQKNKKEPPKKRKTPPEASHPPPRPRPPRPPQPNH 190
Query: 72 AIVKVEKT 79
K +KT
Sbjct: 191QHDKPQKT 214
Score = 32.3 bits (72), Expect = 0.14
Identities = 19/72 (26%), Positives = 26/72 (35%), Gaps = 2/72 (2%)
Frame = +1
Query: 2 TEEQPKKVESES--PSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQP 59
T++Q + E + P PPP P P P + + P + P PQP
Sbjct: 88 TKKQKRAPEKKKNPPRGQPPPPPPPPPQAPPTKPPTRQTPKNNTPPPPPHTPPDPTPPQP 267
Query: 60 SPPESKPVDDSK 71
P KP K
Sbjct: 268 PPQPPKPPHHEK 303
Score = 30.8 bits (68), Expect = 0.41
Identities = 21/80 (26%), Positives = 31/80 (38%), Gaps = 16/80 (20%)
Frame = +1
Query: 3 EEQPKKVESESPSNPPPPAP--------APTESTPAPEAEAKPEPV----VHDAPKEDVA 50
++ P + + P PPP AP P +TP P P+P PK
Sbjct: 118 KKNPPRGQPPPPPPPPPQAPPTKPPTRQTPKNNTPPPPPHTPPDPTPPQPPPQPPKPPHH 297
Query: 51 EEKSIIPQP----SPPESKP 66
E++ P+P PP + P
Sbjct: 298 EKRPRTPEPPGGRPPPPTTP 357
Score = 29.3 bits (64), Expect = 1.2
Identities = 17/65 (26%), Positives = 22/65 (33%), Gaps = 1/65 (1%)
Frame = +1
Query: 3 EEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDV-AEEKSIIPQPSP 61
EE KK + PP P P P +P PK + P P+P
Sbjct: 79 EEATKKQKRAPEKKKNPPRGQPPPPPPPPPQAPPTKPPTRQTPKNNTPPPPPHTPPDPTP 258
Query: 62 PESKP 66
P+ P
Sbjct: 259 PQPPP 273
Score = 28.9 bits (63), Expect = 1.6
Identities = 13/53 (24%), Positives = 23/53 (42%)
Frame = +3
Query: 14 PSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKP 66
P +PP P+P T ++ P H K+ +++ P+P+ P P
Sbjct: 3 PHDPPTPSPQATPQRKGHPSQKPPTGRSHKKTKKSPRKKEKPPPRPATPPPAP 161
Score = 28.5 bits (62), Expect = 2.0
Identities = 20/69 (28%), Positives = 26/69 (36%), Gaps = 8/69 (11%)
Frame = +3
Query: 6 PKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAP--------KEDVAEEKSIIP 57
PKK PP A A E T + EA P P K+ ++K P
Sbjct: 663 PKKTARGGEERPPAGARARREKTRGDQQEAAPAGRPRKKPRGAREREGKKKNPQKKPPKP 842
Query: 58 QPSPPESKP 66
Q PP++ P
Sbjct: 843 QQQPPKNPP 869
Score = 26.9 bits (58), Expect = 5.9
Identities = 11/29 (37%), Positives = 17/29 (57%)
Frame = +3
Query: 3 EEQPKKVESESPSNPPPPAPAPTESTPAP 31
+++P K + + P NPP PT+ TP P
Sbjct: 822 QKKPPKPQQQPPKNPP-----PTKKTPPP 893
Score = 26.6 bits (57), Expect = 7.7
Identities = 16/58 (27%), Positives = 23/58 (39%), Gaps = 9/58 (15%)
Frame = +2
Query: 13 SPSNPPPPAPAPTESTPAPEAEAKPE---------PVVHDAPKEDVAEEKSIIPQPSP 61
+P+ PPPP A TP P+ +K P H P++ P P+P
Sbjct: 407 APAPPPPPPKAREGRTPKPDPGSKENHNRTPGKEGPATHGPPEKKT-------PPPTP 559
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.302 0.122 0.325
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,587,174
Number of Sequences: 36976
Number of extensions: 104123
Number of successful extensions: 4654
Number of sequences better than 10.0: 457
Number of HSP's better than 10.0 without gapping: 1822
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3094
length of query: 215
length of database: 9,014,727
effective HSP length: 92
effective length of query: 123
effective length of database: 5,612,935
effective search space: 690391005
effective search space used: 690391005
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0148.17


