

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0147b.5
(569 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
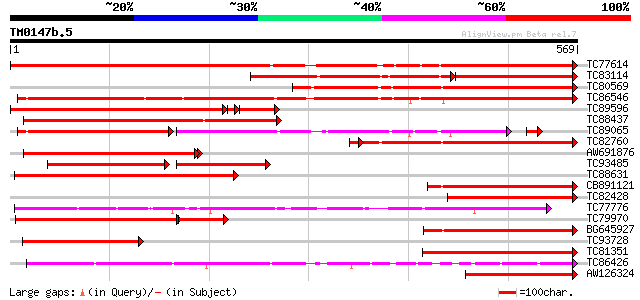
Score E
Sequences producing significant alignments: (bits) Value
TC77614 similar to PIR|JC5229|JC5229 laccase (EC 1.10.3.2) precu... 671 0.0
TC83114 similar to GP|1621467|gb|AAB17194.1| laccase {Liriodendr... 328 e-157
TC80569 similar to GP|9757923|dbj|BAB08370.1 laccase (diphenol o... 499 e-141
TC86546 weakly similar to PIR|T45944|T45944 laccase-like protein... 498 e-141
TC89596 similar to GP|9757923|dbj|BAB08370.1 laccase (diphenol o... 419 e-135
TC88437 similar to GP|1621463|gb|AAB17192.1| laccase {Liriodendr... 445 e-125
TC89065 weakly similar to GP|6478936|gb|AAF14041.1| putative lac... 258 e-118
TC82760 similar to GP|3805964|emb|CAA74105.1 laccase {Populus ba... 353 2e-98
AW691876 similar to GP|3805964|emb laccase {Populus balsamifera ... 318 1e-86
TC93485 similar to GP|21552583|gb|AAM54731.1 diphenol oxidase la... 184 1e-74
TC88631 weakly similar to PIR|T00579|T00579 probable laccase [im... 265 5e-71
CB891121 similar to GP|1621467|gb| laccase {Liriodendron tulipif... 264 8e-71
TC82428 similar to GP|1621461|gb|AAB17191.1| laccase {Liriodendr... 239 2e-63
TC77776 L-ascorbate oxidase 239 3e-63
TC79970 similar to GP|21552583|gb|AAM54731.1 diphenol oxidase la... 199 4e-63
BG645927 similar to GP|11071902|em laccase {Populus balsamifera ... 224 7e-59
TC93728 similar to GP|1621467|gb|AAB17194.1| laccase {Liriodendr... 216 2e-56
TC81351 similar to PIR|T00579|T00579 probable laccase [imported]... 194 8e-50
TC86426 similar to PIR|T07634|T07634 pollen-specific protein hom... 191 5e-49
AW126324 similar to GP|20161818|db putative laccase {Oryza sativ... 191 6e-49
>TC77614 similar to PIR|JC5229|JC5229 laccase (EC 1.10.3.2) precursor -
common tobacco, partial (96%)
Length = 2063
Score = 671 bits (1731), Expect = 0.0
Identities = 321/569 (56%), Positives = 405/569 (70%)
Frame = +2
Query: 1 MITFCIFELALAGTTRHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKV 60
++ CIF + RHY+F + +N +RLC SK++VTVNG+FPGP + ARE D +++KV
Sbjct: 80 LLICCIFPALVECKVRHYKFHVVAKNTSRLCSSKAIVTVNGKFPGPTLYAREDDTVIVKV 259
Query: 61 VNHVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHIS 120
N V NNI+IHWHGIRQL++GWADGPAY+TQCPIQ G SY YN+TI GQRGTL WHAH++
Sbjct: 260 RNQVNNNITIHWHGIRQLRTGWADGPAYITQCPIQPGHSYTYNFTITGQRGTLLWHAHVN 439
Query: 121 WLRATLYGPLIILPKHNAQYPFVKPHKEVPIIFGEWWNADTEAVITQALQTGGGPNVSDA 180
WLR+T++G ++ILPK YPF KP E+ ++ GEWW +DTEAVI +AL++G PNVSDA
Sbjct: 440 WLRSTVHGAIVILPKKGVPYPFPKPDDELVLVLGEWWKSDTEAVINEALKSGLAPNVSDA 619
Query: 181 YTINGLPGPLYNCSQKDTFRLKVKPGKIYLLRLINAALNDELFFSIANHTLTVVEADAGY 240
+TINGLPG + NCS +D ++L V+ GK YLLR+INAALN+ELFF IA H LTVVE DA Y
Sbjct: 620 HTINGLPGTVANCSTQDVYKLPVESGKTYLLRIINAALNEELFFKIAGHKLTVVEVDATY 799
Query: 241 VKPFVTNTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGLGTFDNSTVAGILEYKIQ 300
KPF TI+I PGQTTNVLL K++ + + + A P++ DN T L Y
Sbjct: 800 TKPFQIETIVIAPGQTTNVLL--KANQKSGKYLVAASPFMDAPVAVDNLTATATLHY--- 964
Query: 301 HNTTHHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLRSLASARFPANVPQKVDKHFLF 360
S L P F P N T A F N L+ L S ++P NVP K+D F
Sbjct: 965 -------SGTTLTNTPTFLTTPPPTNATQIANNFLNSLKGLNSKKYPVNVPLKIDHSLFF 1123
Query: 361 TVGLGTNPCKNKSNQTCQGPNNAAMFSASVNNVSFILPTTALLQAHFFGKSNGVYTPDFP 420
TVGLG NPC +C+ N + + A++NNV+F++PTTALLQAH+F N V+T DFP
Sbjct: 1124TVGLGVNPC-----PSCKAGNGSRVV-AAINNVTFVMPTTALLQAHYFNIKN-VFTADFP 1282
Query: 421 TSPLHPFNYTGTTTPNNTNVSNGTKVVVLPFNTSVELVMQDTSILGAESHPLHLHGFNFF 480
+P H +N+TG P N N ++GTK+ L FN +V+LVMQDT I+ ESHP+HLHGFNFF
Sbjct: 1283PNPPHIYNFTG-AGPKNLNTTSGTKLYKLSFNDTVQLVMQDTGIIAPESHPVHLHGFNFF 1459
Query: 481 VVGQGFGNFDPNKDPSKFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTS 540
VVG+G GN+D D KFNLVDPVERNTVGVP+GGWVAIRF ADNPGVWFMHCHLEVHT+
Sbjct: 1460VVGRGVGNYDSKNDSKKFNLVDPVERNTVGVPAGGWVAIRFRADNPGVWFMHCHLEVHTT 1639
Query: 541 WGLRMAWVVLDGKQPNQKLLPPPTDLPKC 569
WGL+MA++V +GK P Q ++ PP DLPKC
Sbjct: 1640WGLKMAFLVDNGKGPKQSVIAPPKDLPKC 1726
>TC83114 similar to GP|1621467|gb|AAB17194.1| laccase {Liriodendron
tulipifera}, partial (54%)
Length = 1216
Score = 328 bits (840), Expect(2) = e-157
Identities = 163/206 (79%), Positives = 179/206 (86%)
Frame = +1
Query: 242 KPFVTNTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGLGTFDNSTVAGILEYKIQH 301
KPFVTNTILI PGQTTNVLLKTK HYPNATF M ARPY TG GTFDNSTVAGI+EY+I
Sbjct: 4 KPFVTNTILIAPGQTTNVLLKTKPHYPNATFLMLARPYATGQGTFDNSTVAGIIEYEIPF 183
Query: 302 NTTHHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLRSLASARFPANVPQKVDKHFLFT 361
NT H NS+ LKKLPL KP+LP LNDTSFAT F+NKL SLA+A+FPANVPQKVDKHF FT
Sbjct: 184 NTHHSNSS--LKKLPLLKPILPQLNDTSFATNFTNKLHSLANAQFPANVPQKVDKHFFFT 357
Query: 362 VGLGTNPCKNKSNQTCQGPNNAAMFSASVNNVSFILPTTALLQAHFFGKSNGVYTPDFPT 421
VGLGTNPC+NK NQTCQGP N MF+ASVNNVSFI+PTTALLQ HFFG++NG+YT DFP+
Sbjct: 358 VGLGTNPCQNK-NQTCQGP-NGTMFAASVNNVSFIMPTTALLQTHFFGQNNGIYTTDFPS 531
Query: 422 SPLHPFNYTGTTTPNNTNVSNGTKVV 447
P++PFNYTG T PNNT VSNGTKVV
Sbjct: 532 KPMNPFNYTG-TPPNNTMVSNGTKVV 606
Score = 247 bits (631), Expect(2) = e-157
Identities = 110/122 (90%), Positives = 115/122 (94%)
Frame = +2
Query: 448 VLPFNTSVELVMQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSKFNLVDPVERN 507
VLPFNTSVELV+QDTSILG ESHPLHLHGFNFFVVGQGFGNFD N DP FNLVDPVERN
Sbjct: 608 VLPFNTSVELVLQDTSILGVESHPLHLHGFNFFVVGQGFGNFDSNSDPQNFNLVDPVERN 787
Query: 508 TVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLRMAWVVLDGKQPNQKLLPPPTDLP 567
TVGVPSGGWVAIRFLADNPGVWFMHCHLE+HTSWGL+MAW+VLDGK PNQK+LPPP DLP
Sbjct: 788 TVGVPSGGWVAIRFLADNPGVWFMHCHLEIHTSWGLKMAWIVLDGKLPNQKVLPPPVDLP 967
Query: 568 KC 569
KC
Sbjct: 968 KC 973
>TC80569 similar to GP|9757923|dbj|BAB08370.1 laccase (diphenol oxidase)
{Arabidopsis thaliana}, partial (46%)
Length = 1117
Score = 499 bits (1284), Expect = e-141
Identities = 242/286 (84%), Positives = 257/286 (89%)
Frame = +1
Query: 284 GTFDNSTVAGILEYKIQHNTTHHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLRSLAS 343
GTFDN+TVAGILEY+I NT H+SA LKK+PLFKP LPALNDTSFATKFS KLRSLAS
Sbjct: 4 GTFDNTTVAGILEYEIPSNT--HHSASSLKKIPLFKPTLPALNDTSFATKFSKKLRSLAS 177
Query: 344 ARFPANVPQKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNAAMFSASVNNVSFILPTTALL 403
+FPANVPQKVDKHF FTVGLGTNPC+ SNQTCQGPN MF+ASVNNVSF +PTTALL
Sbjct: 178 PQFPANVPQKVDKHFFFTVGLGTNPCQ--SNQTCQGPNGT-MFAASVNNVSFTMPTTALL 348
Query: 404 QAHFFGKSNGVYTPDFPTSPLHPFNYTGTTTPNNTNVSNGTKVVVLPFNTSVELVMQDTS 463
Q+HF G+S GVY P FP+SPLHPFNYTGT PNNT VSNGTKV VLPFNTSVELVMQDTS
Sbjct: 349 QSHFTGQSRGVYAPYFPSSPLHPFNYTGTP-PNNTMVSNGTKVTVLPFNTSVELVMQDTS 525
Query: 464 ILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSKFNLVDPVERNTVGVPSGGWVAIRFLA 523
ILGAESHPLHLHGFNFFVVGQGFGN+D NKDP FNLVDPVERNT+GVPSGGWVAIRFLA
Sbjct: 526 ILGAESHPLHLHGFNFFVVGQGFGNYDSNKDPKNFNLVDPVERNTIGVPSGGWVAIRFLA 705
Query: 524 DNPGVWFMHCHLEVHTSWGLRMAWVVLDGKQPNQKLLPPPTDLPKC 569
DNPGVWFMHCHLEVHTSWGL+MAW+VLDGK PNQKLLPPP DLPKC
Sbjct: 706 DNPGVWFMHCHLEVHTSWGLKMAWIVLDGKLPNQKLLPPPADLPKC 843
>TC86546 weakly similar to PIR|T45944|T45944 laccase-like protein -
Arabidopsis thaliana, partial (64%)
Length = 2066
Score = 498 bits (1282), Expect = e-141
Identities = 251/569 (44%), Positives = 358/569 (62%), Gaps = 8/569 (1%)
Frame = +3
Query: 9 LALAGTTRHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVNHVQNNI 68
+A A T H F + + + RLC+ + +VTVNG +PGP++ R+GD +++ V+N+ NI
Sbjct: 84 MASAATVEH-TFLVENKTIKRLCNEQVIVTVNGLYPGPKLEVRDGDSVIVHVINNSPYNI 260
Query: 69 SIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRATLYG 128
+IHWHG+ QL S W+DGP Y+TQC I+ + Y + + Q GTL+WHAH S +RAT++G
Sbjct: 261 TIHWHGVFQLYSAWSDGPEYITQCSIRPENKFTYKFNVTQQEGTLWWHAHASVVRATVHG 440
Query: 129 PLIILPKHNAQYPFVKPHKEVPIIFGEWWNADTEAVITQALQTGGGPNVSDAYTINGLPG 188
+II P+ + ++PF KP+KEVPII G+W++ + E +I + L+TG SDA+TING PG
Sbjct: 441 AIIIQPR-SGRFPFPKPYKEVPIILGDWYDGNVEEIIQKELETGD-KIASDAFTINGFPG 614
Query: 189 PLYNCSQKDTFRLKVKPGKIYLLRLINAALNDELFFSIANHTLTVVEADAGYVKPFVTNT 248
L+NCS+ ++LKVK GK YL R++NAAL + LFF IA+H TVV DA Y +P+ T+
Sbjct: 615 DLFNCSKNQMYKLKVKQGKTYLFRMVNAALANNLFFKIADHKFTVVAMDAAYTEPYTTDI 794
Query: 249 ILITPGQTTNVLLKTKSHYPNATFFMTARPYVTG--LGTFDNSTVAGILEYKIQHNTTHH 306
I+I GQ+ ++L P +++M A PYV G +G FDNST ++ Y+
Sbjct: 795 IVIAAGQSADILFTADQ--PKGSYYMAASPYVVGEPVGLFDNSTTRAVVFYE-------G 947
Query: 307 NSAVKLKKLPLFKPLLPALNDTSFATKFSNKLRSLASARFPANVPQKVDKHFLFTVGLGT 366
+K K + P LP N+T A KF + + L VP +VD+H T+ +G
Sbjct: 948 YKKLKTKHIVPLMPALPLHNNTPIAHKFFSNITGLVGGPNWVPVPLEVDEHMYITINMGL 1127
Query: 367 NPCKNKSNQTCQGPNNAAMFSASVNNVSFILPTT---ALLQAHFFGKSNGVYTPDFPTSP 423
PC N C GP F++S+NN SF+LP ++++A+F+ S G+YT DFP +P
Sbjct: 1128VPCP--VNAKCTGPLGQK-FASSMNNESFLLPVGKGYSIMEAYFYNVS-GIYTTDFPDNP 1295
Query: 424 LHPFNYTGTTT---PNNTNVSNGTKVVVLPFNTSVELVMQDTSILGAESHPLHLHGFNFF 480
F++ PN T TKV +N++VE+V Q+T+IL A+SHP+HLHG NF
Sbjct: 1296PKFFDFVNPKIFLDPNVTFTPKSTKVKQFKYNSTVEIVFQNTAILNAQSHPMHLHGMNFH 1475
Query: 481 VVGQGFGNFDPNKDPSKFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTS 540
V+ Q FG F+P D K+NLV+P RNTV VP GGW AIRF +NPGVWF+HCH++ H
Sbjct: 1476VLAQDFGIFNPTTDKLKYNLVNPSIRNTVAVPVGGWAAIRFRTNNPGVWFLHCHVDDHNL 1655
Query: 541 WGLRMAWVVLDGKQPNQKLLPPPTDLPKC 569
WGL A++V +G P+ L PPP DLPKC
Sbjct: 1656WGLVTAFIVENGPTPSTSLGPPPADLPKC 1742
>TC89596 similar to GP|9757923|dbj|BAB08370.1 laccase (diphenol oxidase)
{Arabidopsis thaliana}, partial (44%)
Length = 921
Score = 419 bits (1078), Expect(3) = e-135
Identities = 194/218 (88%), Positives = 207/218 (93%)
Frame = +2
Query: 1 MITFCIFELALAGTTRHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKV 60
+I FC FE ALAGTTRHYQFDI YQNVTRLCH+K +VTVNGQFPGPRI+AREGDRL+IKV
Sbjct: 110 LINFCAFEFALAGTTRHYQFDIGYQNVTRLCHNKRMVTVNGQFPGPRIMAREGDRLVIKV 289
Query: 61 VNHVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHIS 120
VN+VQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHIS
Sbjct: 290 VNNVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHIS 469
Query: 121 WLRATLYGPLIILPKHNAQYPFVKPHKEVPIIFGEWWNADTEAVITQALQTGGGPNVSDA 180
WLR+TLYGPLIILPK N YPF KPHKEVP+IFGEW+NADTEA+I QALQTGGGPNVS+A
Sbjct: 470 WLRSTLYGPLIILPKKNVPYPFAKPHKEVPMIFGEWFNADTEAIIAQALQTGGGPNVSEA 649
Query: 181 YTINGLPGPLYNCSQKDTFRLKVKPGKIYLLRLINAAL 218
YTINGLPGPLYNCS+KDTF+LKVKPGK YLLRLINAAL
Sbjct: 650 YTINGLPGPLYNCSKKDTFKLKVKPGKTYLLRLINAAL 763
Score = 73.2 bits (178), Expect(3) = e-135
Identities = 36/40 (90%), Positives = 36/40 (90%)
Frame = +1
Query: 231 LTVVEADAGYVKPFVTNTILITPGQTTNVLLKTKSHYPNA 270
L VVEADA YVKPF TNTILI PGQTTNVLLKTKSHYPNA
Sbjct: 802 LKVVEADAIYVKPFETNTILIAPGQTTNVLLKTKSHYPNA 921
Score = 28.9 bits (63), Expect(3) = e-135
Identities = 11/12 (91%), Positives = 12/12 (99%)
Frame = +3
Query: 219 NDELFFSIANHT 230
NDELFFS+ANHT
Sbjct: 765 NDELFFSVANHT 800
>TC88437 similar to GP|1621463|gb|AAB17192.1| laccase {Liriodendron
tulipifera}, partial (41%)
Length = 984
Score = 445 bits (1145), Expect = e-125
Identities = 203/259 (78%), Positives = 233/259 (89%), Gaps = 1/259 (0%)
Frame = +1
Query: 15 TRHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVNHVQNNISIHWHG 74
TRHY+FDIR NVTRLCH+KS+VTVNG+FPGPRI+ REGDRLL+KVVNHV NNIS+HWHG
Sbjct: 190 TRHYKFDIRLANVTRLCHTKSMVTVNGKFPGPRIVVREGDRLLVKVVNHVPNNISLHWHG 369
Query: 75 IRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRATLYGPLIILP 134
+RQL+SGW+DGP+Y+TQCPIQTGQSYVYN+TI GQRGTLFWHAH SWLRAT+YGPLI+LP
Sbjct: 370 VRQLRSGWSDGPSYITQCPIQTGQSYVYNFTIVGQRGTLFWHAHFSWLRATVYGPLILLP 549
Query: 135 KHNAQYPFVKPHKEVPIIFGEWWNADT-EAVITQALQTGGGPNVSDAYTINGLPGPLYNC 193
+HN YPF KP+KEVPI+FGEWW + + + QTGGGPNVSDAYTINGLPGPLYNC
Sbjct: 550 RHNESYPFQKPYKEVPILFGEWWECRS*SCNLHKLFQTGGGPNVSDAYTINGLPGPLYNC 729
Query: 194 SQKDTFRLKVKPGKIYLLRLINAALNDELFFSIANHTLTVVEADAGYVKPFVTNTILITP 253
S KDT++LKVKPGK YLLRLINAALNDELFFSIANHTL +VEADA Y+KPF +NTI++ P
Sbjct: 730 S-KDTYKLKVKPGKTYLLRLINAALNDELFFSIANHTLIIVEADASYIKPFESNTIILGP 906
Query: 254 GQTTNVLLKTKSHYPNATF 272
GQTTNVLLKTK +YPN+TF
Sbjct: 907 GQTTNVLLKTKPNYPNSTF 963
>TC89065 weakly similar to GP|6478936|gb|AAF14041.1| putative laccase
{Arabidopsis thaliana}, partial (63%)
Length = 1647
Score = 258 bits (658), Expect(3) = e-118
Identities = 149/348 (42%), Positives = 203/348 (57%), Gaps = 12/348 (3%)
Frame = +1
Query: 168 ALQTGGGPNVSDAYTINGLPGPLYNCSQKDTFRLKVKPGKIYLLRLINAALNDELFFSIA 227
A TG P S+A TINGLP LYNCSQ T+++KVK GK YLLR+INAALN++ FF IA
Sbjct: 550 ATDTGNPPEESNASTINGLPSDLYNCSQDQTYKVKVKQGKTYLLRIINAALNEQHFFKIA 729
Query: 228 NHTLTVVEADAGYVKPFVTNTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGLGTFD 287
NH+ TVV DA Y + + T+ +++ PGQT +VLLKT +++M PY +
Sbjct: 730 NHSFTVVAMDAIYTEHYNTDVVVLAPGQTVDVLLKTNQVVD--SYYMVFTPYRSSNVGTS 903
Query: 288 NSTVAGILEYKIQHNTTHHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLRSLASARFP 347
N T G++ Y T K P+ P++P +DT A KF + L +
Sbjct: 904 NITTRGVVVYDGASQT----------KAPIM-PIMPDAHDTPTAHKFYTNVTGLTTGPHW 1050
Query: 348 ANVPQKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNAAMFSASVNNVSFILPT---TALLQ 404
VP+KVD+H T G+G C N C N + SA++NN SF+LP +LL+
Sbjct: 1051VPVPRKVDEHMFITFGIGLEQCINPGPGRCVVLN--SRLSANMNNESFVLPKGRGFSLLE 1224
Query: 405 AHFFGKSNGVYTPDFPTSPLHPFNYTGTTTPNNTNVS---------NGTKVVVLPFNTSV 455
A F+ +GVYT DFP P FNY T P+ NV+ TKV L FN++V
Sbjct: 1225A-FYKNISGVYTKDFPNQPPFEFNY---TDPSLANVNPSEPLAFAPKSTKVKTLKFNSTV 1392
Query: 456 ELVMQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSKFNLVDP 503
E+V+Q+T+ILG E+HP+H+HGFNF V+ QGFGN++ +D KFN V+P
Sbjct: 1393EIVLQNTAILGTENHPIHIHGFNFHVLAQGFGNYNATRDEPKFNFVNP 1536
Score = 173 bits (438), Expect(3) = e-118
Identities = 80/156 (51%), Positives = 104/156 (66%)
Frame = +3
Query: 9 LALAGTTRHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVNHVQNNI 68
LA A H F++ + RLC + + VNG PGP I AREGD ++I V N N+
Sbjct: 75 LASAAIVEH-TFNVEDFTIQRLCLPQVITAVNGTLPGPTINAREGDTVIIHVFNKSPYNL 251
Query: 69 SIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRATLYG 128
++HWHGI Q + W+DGP YVTQCPI +G SY Y + + GQ GTL+WHAH S+LRAT++G
Sbjct: 252 TLHWHGIIQFLTPWSDGPEYVTQCPIPSGGSYTYQFNLTGQEGTLWWHAHSSFLRATVHG 431
Query: 129 PLIILPKHNAQYPFVKPHKEVPIIFGEWWNADTEAV 164
LII P+ YPF K ++EVPI+ GEWWNA+ E V
Sbjct: 432 ALIIKPRLGRSYPFPKVYQEVPILLGEWWNANVEEV 539
Score = 33.9 bits (76), Expect(3) = e-118
Identities = 10/16 (62%), Positives = 14/16 (87%)
Frame = +3
Query: 519 IRFLADNPGVWFMHCH 534
+RF A+NPG+W +HCH
Sbjct: 1584 VRFQANNPGIWLVHCH 1631
>TC82760 similar to GP|3805964|emb|CAA74105.1 laccase {Populus balsamifera
subsp. trichocarpa}, partial (34%)
Length = 970
Score = 353 bits (905), Expect(2) = 2e-98
Identities = 165/220 (75%), Positives = 190/220 (86%), Gaps = 2/220 (0%)
Frame = +3
Query: 352 QKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNAAMFSASVNNVSFILPTTALLQAHFFGKS 411
+K+ F FTVGLGT PC N TCQGPNN F+ASVNN SF+LP+ +++QA++FGKS
Sbjct: 33 KKLTIKFFFTVGLGTFPCPK--NSTCQGPNNNTKFAASVNNFSFVLPSVSIMQAYYFGKS 206
Query: 412 N--GVYTPDFPTSPLHPFNYTGTTTPNNTNVSNGTKVVVLPFNTSVELVMQDTSILGAES 469
N GVY DFP +PL+PFNYTGT+ PNNT V+N TK+VVL FNTSVELV+QDTSILGAES
Sbjct: 207 NSNGVYKTDFPETPLNPFNYTGTS-PNNTMVNNDTKLVVLNFNTSVELVLQDTSILGAES 383
Query: 470 HPLHLHGFNFFVVGQGFGNFDPNKDPSKFNLVDPVERNTVGVPSGGWVAIRFLADNPGVW 529
HPLHLHG++FFVVGQGFGN+D NKDP+KFNLVDPVERNTVGVP+GGWVAIRF ADNPGVW
Sbjct: 384 HPLHLHGYDFFVVGQGFGNYDANKDPAKFNLVDPVERNTVGVPAGGWVAIRFFADNPGVW 563
Query: 530 FMHCHLEVHTSWGLRMAWVVLDGKQPNQKLLPPPTDLPKC 569
FMHCHL++HTSWGLRMAW+VLDG NQKL PPP+DLPKC
Sbjct: 564 FMHCHLDIHTSWGLRMAWLVLDGPDSNQKLQPPPSDLPKC 683
Score = 25.0 bits (53), Expect(2) = 2e-98
Identities = 9/14 (64%), Positives = 13/14 (92%)
Frame = +2
Query: 342 ASARFPANVPQKVD 355
A+++FP NVP+KVD
Sbjct: 2 ANSKFPINVPKKVD 43
>AW691876 similar to GP|3805964|emb laccase {Populus balsamifera subsp.
trichocarpa}, partial (30%)
Length = 659
Score = 318 bits (815), Expect(2) = 1e-86
Identities = 136/176 (77%), Positives = 161/176 (91%)
Frame = +1
Query: 15 TRHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVNHVQNNISIHWHG 74
TRHY F+I Y+NVTRLCH++++++VNG+FPGPR++AREGDR+L+KVVNH+ NN++IHWHG
Sbjct: 103 TRHYTFNIEYKNVTRLCHTRTILSVNGKFPGPRLVAREGDRVLVKVVNHISNNVTIHWHG 282
Query: 75 IRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRATLYGPLIILP 134
IRQ +GW+DGPAYVTQCPIQT Q+Y YN+TI GQRGTLFWHAHISWLRATLYGP+IILP
Sbjct: 283 IRQKTTGWSDGPAYVTQCPIQTNQTYTYNFTITGQRGTLFWHAHISWLRATLYGPIIILP 462
Query: 135 KHNAQYPFVKPHKEVPIIFGEWWNADTEAVITQALQTGGGPNVSDAYTINGLPGPL 190
KHN YPF KPHKE+PI+FGEW+N D EAVI QALQTGGGPNVSDAYTINGLPG +
Sbjct: 463 KHNESYPFQKPHKEIPILFGEWFNVDPEAVINQALQTGGGPNVSDAYTINGLPGTI 630
Score = 20.8 bits (42), Expect(2) = 1e-86
Identities = 6/7 (85%), Positives = 6/7 (85%)
Frame = +2
Query: 187 PGPLYNC 193
PGP YNC
Sbjct: 620 PGPFYNC 640
>TC93485 similar to GP|21552583|gb|AAM54731.1 diphenol oxidase laccase
{Glycine max}, partial (40%)
Length = 740
Score = 184 bits (467), Expect(2) = 1e-74
Identities = 75/122 (61%), Positives = 96/122 (78%)
Frame = +3
Query: 39 VNGQFPGPRIIAREGDRLLIKVVNHVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQ 98
VNGQFPGP + GD L++KV+N + N++IHWHG+RQ+++GWADGP +VTQCPI+ G+
Sbjct: 48 VNGQFPGPTLEINNGDTLVVKVINKARYNVTIHWHGVRQIRTGWADGPEFVTQCPIRPGE 227
Query: 99 SYVYNYTIKGQRGTLFWHAHISWLRATLYGPLIILPKHNAQYPFVKPHKEVPIIFGEWWN 158
SY Y +TI GQ GTL+WHAH SWLRAT+YG LII PK YPF KP +E PI+ GEWW+
Sbjct: 228 SYTYRFTINGQEGTLWWHAHSSWLRATVYGALIIHPKEGDAYPFTKPKRETPILLGEWWD 407
Query: 159 AD 160
A+
Sbjct: 408 AN 413
Score = 114 bits (285), Expect(2) = 1e-74
Identities = 55/94 (58%), Positives = 67/94 (70%)
Frame = +2
Query: 168 ALQTGGGPNVSDAYTINGLPGPLYNCSQKDTFRLKVKPGKIYLLRLINAALNDELFFSIA 227
A QTG PN+SDAYTING PG LY CS K T + + G+ L+R+INAALN L F+IA
Sbjct: 437 ATQTGAAPNISDAYTINGQPGDLYKCSSKGTTIVPIXSGETNLIRVINAALNQPLXFTIA 616
Query: 228 NHTLTVVEADAGYVKPFVTNTILITPGQTTNVLL 261
NH LT ADA YVKPF TN +++ P QTT+VL+
Sbjct: 617 NHKLTXXGADASYVKPFTTNVLMLGPDQTTDVLI 718
>TC88631 weakly similar to PIR|T00579|T00579 probable laccase [imported] -
Arabidopsis thaliana, partial (38%)
Length = 779
Score = 265 bits (676), Expect = 5e-71
Identities = 126/224 (56%), Positives = 157/224 (69%)
Frame = +3
Query: 6 IFELALAGTTRHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVNHVQ 65
I A A ++QF I+ V RLC ++ ++TVNGQFPGP I AR+GD ++IKV N
Sbjct: 69 IVSFASAAENHYHQFVIQTATVKRLCKTRRILTVNGQFPGPTIEARDGDSMVIKVTNAGP 248
Query: 66 NNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRAT 125
NISIHWHG R L++ WADGP+YVTQCPIQ G SY Y +TI+ Q GTL+WHAH +LRAT
Sbjct: 249 YNISIHWHGFRMLRNPWADGPSYVTQCPIQPGGSYTYRFTIQNQEGTLWWHAHTGFLRAT 428
Query: 126 LYGPLIILPKHNAQYPFVKPHKEVPIIFGEWWNADTEAVITQALQTGGGPNVSDAYTING 185
+YG II PK + YPF P +E PI+ GEW++ D A++ Q TG PNVS AYT+NG
Sbjct: 429 VYGAFIIYPKMGSPYPFSMPTREFPILLGEWFDRDPMALLRQTQFTGAPPNVSVAYTMNG 608
Query: 186 LPGPLYNCSQKDTFRLKVKPGKIYLLRLINAALNDELFFSIANH 229
PG LY CS + T R +V G+ LLR+IN+ALN ELFFSIANH
Sbjct: 609 QPGDLYRCSSQGTVRFQVYAGETILLRIINSALNQELFFSIANH 740
>CB891121 similar to GP|1621467|gb| laccase {Liriodendron tulipifera},
partial (25%)
Length = 706
Score = 264 bits (674), Expect = 8e-71
Identities = 120/150 (80%), Positives = 133/150 (88%)
Frame = -3
Query: 420 PTSPLHPFNYTGTTTPNNTNVSNGTKVVVLPFNTSVELVMQDTSILGAESHPLHLHGFNF 479
P++P FNYTGT P N V +GTKV VLP+NT VELV+QDTSILGAESHPLHLHGFNF
Sbjct: 704 PSNPPVKFNYTGTP-PKNIMVKSGTKVAVLPYNTKVELVLQDTSILGAESHPLHLHGFNF 528
Query: 480 FVVGQGFGNFDPNKDPSKFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHT 539
F+VGQG GNFDP KDP+KFNLVDPVERNT GVP+GGWVA+RFLADNPGVWFMHCHLEVHT
Sbjct: 527 FIVGQGNGNFDPKKDPAKFNLVDPVERNTAGVPAGGWVALRFLADNPGVWFMHCHLEVHT 348
Query: 540 SWGLRMAWVVLDGKQPNQKLLPPPTDLPKC 569
SWGL+MAW+V DGK+ NQKL PPP+DLPKC
Sbjct: 347 SWGLKMAWIVQDGKRRNQKLPPPPSDLPKC 258
>TC82428 similar to GP|1621461|gb|AAB17191.1| laccase {Liriodendron
tulipifera}, partial (22%)
Length = 707
Score = 239 bits (610), Expect = 2e-63
Identities = 106/130 (81%), Positives = 118/130 (90%)
Frame = +1
Query: 440 VSNGTKVVVLPFNTSVELVMQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSKFN 499
VSNGTK VV+P+NT V++++QDTSILGAESHPLHLHGFNFFVVGQGFGNF+ + DP+KFN
Sbjct: 4 VSNGTKTVVIPYNTRVQVILQDTSILGAESHPLHLHGFNFFVVGQGFGNFNASSDPAKFN 183
Query: 500 LVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLRMAWVVLDGKQPNQKL 559
LVDPVERNTV VPSGGWVAIRFLADNPGVW MHCH +VH SWGLRMAW+V DGK P+QKL
Sbjct: 184 LVDPVERNTVAVPSGGWVAIRFLADNPGVWLMHCHFDVHLSWGLRMAWIVEDGKLPDQKL 363
Query: 560 LPPPTDLPKC 569
PPP DLPKC
Sbjct: 364 PPPPKDLPKC 393
>TC77776 L-ascorbate oxidase
Length = 2066
Score = 239 bits (609), Expect = 3e-63
Identities = 184/571 (32%), Positives = 260/571 (45%), Gaps = 33/571 (5%)
Frame = +3
Query: 6 IFELALAGTTRHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVN--H 63
+FEL+LA HY+FD+ Y C ++ +NGQFPGP I A GD L+I + N H
Sbjct: 93 LFELSLAKGKSHYKFDVEYIYKKPDCKEHVVMGINGQFPGPTIRAEVGDTLVIDLTNKLH 272
Query: 64 VQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLR 123
+ + IHWHGIRQ + WADG A ++QC I G+++ Y + + + GT F+H H R
Sbjct: 273 TEGTV-IHWHGIRQFGTPWADGTAAISQCAINPGETFQYKFKVD-RPGTYFYHGHYGMQR 446
Query: 124 AT-LYGPLII-LPKHNAQYPFVKPHKEVPIIFGEWWNADTE----AVITQALQTGGGPNV 177
A LYG LI+ LPK + PF E ++ + W+ + + + ++ G P
Sbjct: 447 AAGLYGSLIVDLPKSQRE-PF-HYDGEFNLLLSDLWHTSSHEQEVGLSSAPMRWIGEPQ- 617
Query: 178 SDAYTINGLPGPLYNCSQKDTFR-------------------LKVKPGKIYLLRLINAAL 218
+ ING +NCS + L V+P K Y +R+ +
Sbjct: 618 --SLLINGRGQ--FNCSLASKYGSTNLPQCNLKGGEECAPQILHVEPKKTYRIRIASTTS 785
Query: 219 NDELFFSIANHTLTVVEADAGYVKPFVTNTILITPGQTTNVLLKTKSHYPNATFFMTARP 278
L +I+NH L VVEAD YV PF + I I G+T +VLL T P ++++
Sbjct: 786 LASLNLAISNHKLIVVEADGNYVHPFAVDDIDIYSGETYSVLLTTDQD-PKKNYWLSI-- 956
Query: 279 YVTGLGTFDNSTVAG--ILEYKIQHNTTHHNSAVKLKKLPLFKPLLPALNDTSFATKFSN 336
G+ ST IL YK + S P+ P NDT + F+
Sbjct: 957 ---GVRGRKPSTPQALTILNYKPLSASVFPTSP---------PPVTPLWNDTDHSKAFTK 1100
Query: 337 KLRSLASARFPANVPQKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNAAMFSA-SVNNVSF 395
++ S P + H L T N F+ ++NNVS
Sbjct: 1101QIISKMGNPQPPKSSHRTI-HLLNT------------------QNKIGSFTKWAINNVSL 1223
Query: 396 ILPTTALLQAHFFGKSNGVYTPDFPTSPLHPFNYTGTTTPNNTNVSNGTKVVVLPFNTSV 455
LPTT L + F N P P +Y P N N + G V N V
Sbjct: 1224TLPTTPYLGSIKFNLKNTF--DKNPPPERFPMDYDIFKNPVNPNTTTGNGVYTFQLNEVV 1397
Query: 456 ELVMQDTSIL---GAESHPLHLHGFNFFVVGQGFGNFDPNKDPSKFNLVDPVERNTVGVP 512
++++Q+ + L G+E HP HLHG +F+V+G G G F P D FNL RNT +
Sbjct: 1398DVILQNANQLNGNGSEIHPWHLHGHDFWVLGYGEGRFRPGVDERSFNLTRAPLRNTAVIF 1577
Query: 513 SGGWVAIRFLADNPGVWFMHCHLEVHTSWGL 543
GW A+RF ADNPGVW HCH+E H G+
Sbjct: 1578PYGWTALRFKADNPGVWAFHCHIEPHLHMGM 1670
>TC79970 similar to GP|21552583|gb|AAM54731.1 diphenol oxidase laccase
{Glycine max}, partial (36%)
Length = 800
Score = 199 bits (506), Expect(2) = 4e-63
Identities = 87/167 (52%), Positives = 117/167 (69%), Gaps = 1/167 (0%)
Frame = +1
Query: 7 FELALAGTTRH-YQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVNHVQ 65
F LALA H ++F + V RLC + + +TVNGQ+PGP + GD L++KV N +
Sbjct: 151 FALALANPKTHEHEFVVEATPVKRLCKTHNTITVNGQYPGPTLEINNGDTLVVKVTNKAR 330
Query: 66 NNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRAT 125
N++IHWHG+RQ+++GWADGP +VTQCPI+ G SY Y +T+ GQ GTL+WHAH SWLRAT
Sbjct: 331 YNVTIHWHGVRQMRTGWADGPEFVTQCPIRPGGSYTYRFTVNGQEGTLWWHAHSSWLRAT 510
Query: 126 LYGPLIILPKHNAQYPFVKPHKEVPIIFGEWWNADTEAVITQALQTG 172
+YG LII P+ YPF KP+ E I+ GEWW+ + V+ QA +TG
Sbjct: 511 VYGALIIRPREGEPYPFPKPNHETSILLGEWWDGNPINVVRQAQRTG 651
Score = 61.2 bits (147), Expect(2) = 4e-63
Identities = 28/50 (56%), Positives = 35/50 (70%)
Frame = +2
Query: 170 QTGGGPNVSDAYTINGLPGPLYNCSQKDTFRLKVKPGKIYLLRLINAALN 219
+ G PN+SDAYTING PG LY CS K T + + G+ L+R+INAALN
Sbjct: 644 ELGAAPNISDAYTINGQPGDLYKCSTKGTTIIPIHSGETNLVRVINAALN 793
>BG645927 similar to GP|11071902|em laccase {Populus balsamifera subsp.
trichocarpa}, partial (38%)
Length = 574
Score = 224 bits (571), Expect = 7e-59
Identities = 101/154 (65%), Positives = 125/154 (80%)
Frame = -1
Query: 416 TPDFPTSPLHPFNYTGTTTPNNTNVSNGTKVVVLPFNTSVELVMQDTSILGAESHPLHLH 475
T DFP +P +N+TG+ N + GT++ L +N++V+LV+QDT +L E+HP+HLH
Sbjct: 574 TDDFPGNPPVVYNFTGSQV-TNLATTKGTRLYRLAYNSTVQLVLQDTGMLTPENHPIHLH 398
Query: 476 GFNFFVVGQGFGNFDPNKDPSKFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHL 535
GFNFFVVG+G GNFD KD KFNLVDPVERNTVGVP+GGW AIRF ADNPGVWFMHCHL
Sbjct: 397 GFNFFVVGRGQGNFDSKKDVKKFNLVDPVERNTVGVPAGGWTAIRFKADNPGVWFMHCHL 218
Query: 536 EVHTSWGLRMAWVVLDGKQPNQKLLPPPTDLPKC 569
E+HT+WGL+MA+VV +GK PN+ LLPPP+DLPKC
Sbjct: 217 EIHTTWGLKMAFVVDNGKGPNESLLPPPSDLPKC 116
>TC93728 similar to GP|1621467|gb|AAB17194.1| laccase {Liriodendron
tulipifera}, partial (21%)
Length = 514
Score = 216 bits (549), Expect = 2e-56
Identities = 94/121 (77%), Positives = 114/121 (93%)
Frame = +2
Query: 14 TTRHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVNHVQNNISIHWH 73
TTRHY+FDI+ QNVTRLC +KS+VTVNGQFPGPRIIAREGDR+++KVVNHVQ N+SIHWH
Sbjct: 152 TTRHYKFDIKLQNVTRLCQTKSIVTVNGQFPGPRIIAREGDRVVVKVVNHVQYNVSIHWH 331
Query: 74 GIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRATLYGPLIIL 133
GIRQ++S WADGPAY+TQCPI+ GQSYV+ +TI GQRGTL++HAHISWLR+TL+GP++IL
Sbjct: 332 GIRQVRSAWADGPAYITQCPIKPGQSYVHKFTIIGQRGTLWYHAHISWLRSTLHGPIVIL 511
Query: 134 P 134
P
Sbjct: 512 P 514
>TC81351 similar to PIR|T00579|T00579 probable laccase [imported] -
Arabidopsis thaliana, partial (27%)
Length = 765
Score = 194 bits (493), Expect = 8e-50
Identities = 85/155 (54%), Positives = 110/155 (70%)
Frame = +3
Query: 415 YTPDFPTSPLHPFNYTGTTTPNNTNVSNGTKVVVLPFNTSVELVMQDTSILGAESHPLHL 474
+T DFP FNYTG GTK+ L + ++V++V+QDTSI+ E HP+H+
Sbjct: 54 FTTDFPPVLPIQFNYTGNVPRGLWTPRKGTKLFKLKYGSNVQIVLQDTSIVTVEDHPMHV 233
Query: 475 HGFNFFVVGQGFGNFDPNKDPSKFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCH 534
HGF+FFVVG GFGNF+P DP+ FNLVDP RNT+G P GGWVAIRF ADNPG+WF+HCH
Sbjct: 234 HGFHFFVVGSGFGNFNPRTDPATFNLVDPPVRNTIGTPPGGWVAIRFKADNPGIWFLHCH 413
Query: 535 LEVHTSWGLRMAWVVLDGKQPNQKLLPPPTDLPKC 569
++ H +WGL A +V +G P Q ++PPP DLP+C
Sbjct: 414 IDSHLNWGLGTALLVENGVGPLQSVIPPPPDLPQC 518
>TC86426 similar to PIR|T07634|T07634 pollen-specific protein homolog
T1P17.10 - Arabidopsis thaliana, partial (91%)
Length = 2220
Score = 191 bits (486), Expect = 5e-49
Identities = 163/564 (28%), Positives = 252/564 (43%), Gaps = 12/564 (2%)
Frame = +3
Query: 18 YQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVNHVQNNISIHWHGIRQ 77
Y+F++ Y + L + ++ +N QFPGP I + + + V N + N+ IHW G++Q
Sbjct: 261 YEFEVSYITASPLGVPQQVIAINKQFPGPTINVTTNNNVAVNVHNKLDENLLIHWSGVQQ 440
Query: 78 LQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRAT-LYGPLIILPKH 136
+S W DG T CPI ++ Y + +K Q G+ F+ +++ RA +G II +
Sbjct: 441 RRSSWQDG-VLGTNCPIPPKWNWTYQFQVKDQIGSFFYFPSLNFQRAAGGFGGFIINNRP 617
Query: 137 NAQYPFVKPHKEVPIIFGEWWNADTEAVITQALQTGGGPNVSDAYTINGLPGPLYNCSQK 196
PF P +++ + G+W+ + A + +AL G + D ING YN +
Sbjct: 618 VISVPFDTPERDIVVFIGDWYTRNHTA-LRKALDDGKDLGMPDGVLINGKGPYRYNDTLV 794
Query: 197 ----DTFRLKVKPGKIYLLRLINAALNDELFFSIANHTLTVVEADAGYVKPFVTNTILIT 252
D ++ VKPGK Y LR+ N ++ L F I NH L + E + Y ++ I
Sbjct: 795 PEGIDFEQIDVKPGKTYRLRVHNVGISTSLNFRIQNHNLLLAETEGSYTVQQNYTSLDIH 974
Query: 253 PGQTTNVLLKTKSHYPNATFFMTARPYVTGLGTFDNSTVAGILEYKIQHNTTHHNSAVKL 312
GQ+ + L+ T ++ +++ A + T GIL Y NS K
Sbjct: 975 VGQSYSFLV-TMDQNASSDYYIVASARFVNESRWQRVTGVGILHYS--------NSKGKA 1127
Query: 313 KKLPLFKPLLPALNDTSFATKFS-NKLRSL---ASARFPANVPQKVDKHFLFTVGLGTNP 368
+ LP D F FS N+ RS+ SA PQ ++ V
Sbjct: 1128------RGHLPPGPDDQFDKTFSMNQARSIRWNVSASGARPNPQGSFRYGSINV-TEIYV 1286
Query: 369 CKNKSNQTCQGPNNAAMFSASVNNVSFILPTTALLQAHFFGKSNGVYTPDFPTSPLHPFN 428
KNK G A+++ +SF P T + A + K GVY DFPT PL
Sbjct: 1287LKNKPPVKIDGKRR-----ATLSGISFANPATPIRLADHY-KLKGVYKLDFPTKPL---- 1436
Query: 429 YTGTTTPN-NTNVSNGTKVVVLPFNTSVELVMQDTSILGAESHPLHLHGFNFFVVGQGFG 487
T +P T+V NG+ F +E+++Q+ + H HL G+ FFVVG FG
Sbjct: 1437---TGSPRVETSVINGS------FRGFMEIILQNND---TKMHTYHLSGYAFFVVGMDFG 1580
Query: 488 NFDPNKDPSKFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSW--GLRM 545
++ N +N D + R+T V G W A+ DN GVW + E SW G
Sbjct: 1581DWSEN-SRGTYNKWDGIARSTAQVYPGAWTAVLVSLDNVGVW--NLRTENLDSWYLGQET 1751
Query: 546 AWVVLDGKQPNQKLLPPPTDLPKC 569
V++ + N+ LP P + KC
Sbjct: 1752YIRVVNPEPTNKTELPMPDNALKC 1823
>AW126324 similar to GP|20161818|db putative laccase {Oryza sativa (japonica
cultivar-group)}, partial (19%)
Length = 535
Score = 191 bits (485), Expect = 6e-49
Identities = 82/112 (73%), Positives = 96/112 (85%)
Frame = +2
Query: 458 VMQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSKFNLVDPVERNTVGVPSGGWV 517
V+QDT++L ESHP HLHG+NFFVVG G GNFDP KDP+K+NLVDP+ERNTVGVP+GGW
Sbjct: 2 VLQDTNLLTVESHPFHLHGYNFFVVGTGIGNFDPAKDPAKYNLVDPMERNTVGVPTGGWT 181
Query: 518 AIRFLADNPGVWFMHCHLEVHTSWGLRMAWVVLDGKQPNQKLLPPPTDLPKC 569
AIRF A+NPGVWFMHCHLE+HT WGL+ A+VV DG +Q +LPPP DLPKC
Sbjct: 182 AIRFTANNPGVWFMHCHLELHTGWGLKTAFVVEDGPGKDQSVLPPPKDLPKC 337
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,394,269
Number of Sequences: 36976
Number of extensions: 317019
Number of successful extensions: 1696
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 1612
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1635
length of query: 569
length of database: 9,014,727
effective HSP length: 101
effective length of query: 468
effective length of database: 5,280,151
effective search space: 2471110668
effective search space used: 2471110668
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0147b.5


