

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0147b.4
(313 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
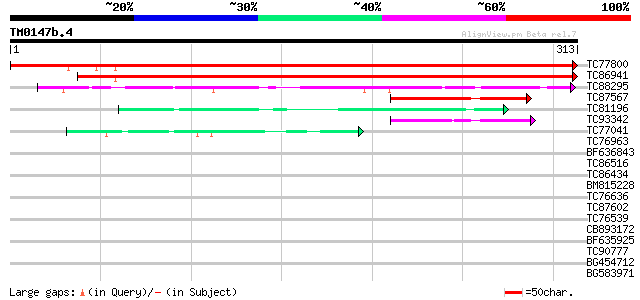
Score E
Sequences producing significant alignments: (bits) Value
TC77800 similar to PIR|T02532|T02532 hypothetical protein At2g37... 494 e-140
TC86941 similar to GP|15294290|gb|AAK95322.1 AT5g02240/T7H20_290... 422 e-119
TC88295 similar to GP|15912295|gb|AAL08281.1 At2g34460/T31E10.20... 79 3e-15
TC87567 similar to GP|17065112|gb|AAL32710.1 Unknown protein {Ar... 59 2e-09
TC81196 similar to PIR|A96751|A96751 hypothetical protein F28P22... 52 2e-07
TC93342 homologue to GP|20339364|gb|AAM19355.1 UOS1 {Pisum sativ... 49 2e-06
TC77041 similar to GP|3716262|emb|CAA03625.1 unnamed protein pro... 41 7e-04
TC76963 GP|3716262|emb|CAA03625.1 unnamed protein product {unide... 38 0.006
BF636843 homologue to PIR|T10683|T106 hypothetical protein F3L17... 36 0.017
TC86516 homologue to GP|18087583|gb|AAL58922.1 AT4g35250/F23E12_... 34 0.064
TC86434 similar to GP|2262100|gb|AAB63608.1| unknown protein {Ar... 30 1.2
BM815228 similar to GP|22136000|gb unknown protein {Arabidopsis ... 30 1.6
TC76636 homologue to GP|6625788|gb|AAF19398.1| dynamin homolog {... 29 2.0
TC87602 similar to GP|21741453|emb|CAD41397. OJ000223_09.9 {Oryz... 29 2.0
TC76539 aquaporin [Medicago truncatula] 29 2.0
CB893172 homologue to PIR|T45642|T45 FtsH metalloproteinase-like... 29 2.0
BF635925 similar to GP|8489806|gb chloroplast protein import com... 29 2.7
TC90777 weakly similar to GP|15810655|gb|AAL07252.1 putative ubi... 28 3.5
BG454712 similar to GP|6469135|emb| putative water channel prote... 28 3.5
BG583971 homologue to GP|6469135|emb putative water channel prot... 28 4.6
>TC77800 similar to PIR|T02532|T02532 hypothetical protein At2g37660
[imported] - Arabidopsis thaliana, partial (73%)
Length = 1379
Score = 494 bits (1272), Expect = e-140
Identities = 259/326 (79%), Positives = 285/326 (86%), Gaps = 13/326 (3%)
Frame = +1
Query: 1 MGMATRVPFVSAT-LSPNQCHKYCLPSSLKLL----TSLQFSHSSSLSLAT------HKR 49
M TRVPFVSAT PNQCHKY L + L TSL+ S S SL + KR
Sbjct: 184 MATTTRVPFVSATTFFPNQCHKYSLVARTINLPVSSTSLRLSSCYSTSLVSLALPRSFKR 363
Query: 50 VRTRTSVV--AMADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIG 107
R SVV AMA+SS+STVLVTGAGGRTG+IVYKKL+ER ++YIARGLVR+EESKQ IG
Sbjct: 364 GGNRRSVVVVAMAESSKSTVLVTGAGGRTGQIVYKKLKERPNEYIARGLVRSEESKQKIG 543
Query: 108 ASDDVYVGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQ 167
A+DDV++GDIRDT S+APAIQGIDALIILTS VPLMKPGF+PT+G+RPEFYFEDGAYPEQ
Sbjct: 544 AADDVFIGDIRDTESLAPAIQGIDALIILTSGVPLMKPGFDPTQGKRPEFYFEDGAYPEQ 723
Query: 168 VDWIGQKNQIDAAKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGI 227
VDWIGQKNQIDAAKAAGVKQIVLVGSMGGTDLN+PLNSLG+GNILVWKRKAEQYLADSGI
Sbjct: 724 VDWIGQKNQIDAAKAAGVKQIVLVGSMGGTDLNHPLNSLGDGNILVWKRKAEQYLADSGI 903
Query: 228 PYTIIRAGGLQDKEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFD 287
PYTIIRAGGLQDKEGG+REL+IGKDDE+LKT+ RTIARPDVAEVC+QALNFEEAQFKAFD
Sbjct: 904 PYTIIRAGGLQDKEGGIRELVIGKDDELLKTDIRTIARPDVAEVCLQALNFEEAQFKAFD 1083
Query: 288 LASKPEGAGTPTRDFKALFSQITTRF 313
LASKPEG G+PT+DFKALFSQITTRF
Sbjct: 1084LASKPEGTGSPTKDFKALFSQITTRF 1161
>TC86941 similar to GP|15294290|gb|AAK95322.1 AT5g02240/T7H20_290
{Arabidopsis thaliana}, partial (98%)
Length = 1121
Score = 422 bits (1085), Expect = e-119
Identities = 211/279 (75%), Positives = 243/279 (86%), Gaps = 3/279 (1%)
Frame = +2
Query: 38 HSSSLSLATHKRVRTRTSVV---AMADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIAR 94
HSSS S +H + +MA SS++TVLVTGAGGRTG+IVYKKL+E+ QYIAR
Sbjct: 29 HSSSSSSVSHFQSIEHCYCFKFNSMAGSSQTTVLVTGAGGRTGQIVYKKLKEKRDQYIAR 208
Query: 95 GLVRTEESKQTIGASDDVYVGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGER 154
GLVR+EESKQ IG +DD+++GDIR+ SI PAIQG DALIILTSAVP MKPGF+PTKG R
Sbjct: 209 GLVRSEESKQKIGGADDIFLGDIRNAESIVPAIQGTDALIILTSAVPQMKPGFDPTKGGR 388
Query: 155 PEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVW 214
PEFYF+DGAYPEQVDWIGQKNQIDAAKAAGVK IVLVGSMGGT+ N+PLNSLGNGNILVW
Sbjct: 389 PEFYFDDGAYPEQVDWIGQKNQIDAAKAAGVKHIVLVGSMGGTNPNHPLNSLGNGNILVW 568
Query: 215 KRKAEQYLADSGIPYTIIRAGGLQDKEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQ 274
KRKAE+YL++SG+PYTIIR GGL DKEGGVRELI+GKDDE+L+TET+TI R DVAEVC+Q
Sbjct: 569 KRKAEEYLSNSGVPYTIIRPGGLLDKEGGVRELIVGKDDELLQTETKTIPRADVAEVCVQ 748
Query: 275 ALNFEEAQFKAFDLASKPEGAGTPTRDFKALFSQITTRF 313
LN+EE + KAFDLASKPEGAG PT+DFKALFSQ+T+RF
Sbjct: 749 VLNYEETKLKAFDLASKPEGAGEPTKDFKALFSQLTSRF 865
>TC88295 similar to GP|15912295|gb|AAL08281.1 At2g34460/T31E10.20
{Arabidopsis thaliana}, partial (93%)
Length = 967
Score = 78.6 bits (192), Expect = 3e-15
Identities = 82/310 (26%), Positives = 142/310 (45%), Gaps = 13/310 (4%)
Frame = +1
Query: 16 PNQCHKYCLPSSL--KLLTSLQFSHSSSLSLATHKRVRTRTSVVAMADSSRSTVLVTGAG 73
P+ H + PSS K LT + F+ + T + V S+ + V V GA
Sbjct: 58 PSHTHHFTAPSSFRTKSLTIINFAKMEGSEI-TQQAVDDDLSL-------KKKVFVAGAT 213
Query: 74 GRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDD---VYVGDIRDTGSIAPAI-QG 129
G TGK + ++L + + G+ +++K ++ A+ V V + +A AI
Sbjct: 214 GSTGKRIVEQLLAKGFA-VKAGVRDLDKAKTSLSANPSLQFVKVDVTEGSDKLAEAIGDD 390
Query: 130 IDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIV 189
+A++ T +PG+ D P +VD G N ++A + V + +
Sbjct: 391 TEAVVCATG----FRPGW-------------DLLAPWKVDNFGTVNLVEACRKVNVNRFI 519
Query: 190 LVGSM-----GGTDLNNPLNSLGN--GNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEG 242
L+ S+ L NP N G LV K +AE ++ SGI YTIIR GGL++ +
Sbjct: 520 LISSILVNGAAMGQLLNPAYIFLNVFGLTLVAKLQAENHIRKSGINYTIIRPGGLKN-DP 696
Query: 243 GVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDF 302
+++ +D + + +I+R VAEV +++L + EA +K ++ ++P+ P R +
Sbjct: 697 PTGNVVMEPEDTLYE---GSISRDQVAEVAVESLAYPEASYKVVEIVARPD---APKRAY 858
Query: 303 KALFSQITTR 312
LF I R
Sbjct: 859 HDLFGSIVQR 888
>TC87567 similar to GP|17065112|gb|AAL32710.1 Unknown protein {Arabidopsis
thaliana}, partial (82%)
Length = 2117
Score = 59.3 bits (142), Expect = 2e-09
Identities = 33/78 (42%), Positives = 49/78 (62%)
Frame = +2
Query: 211 ILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRELIIGKDDEILKTETRTIARPDVAE 270
+L KR E L SG+ YTI+R G LQ++ GG R LI + D I +R I+ DVA+
Sbjct: 1529 VLKAKRAGEDSLRRSGLGYTIVRPGPLQEEPGGQRALIFDQGDRI----SRGISCADVAD 1696
Query: 271 VCIQALNFEEAQFKAFDL 288
+C++AL+ A+ K+FD+
Sbjct: 1697 ICVKALHDSTARNKSFDV 1750
>TC81196 similar to PIR|A96751|A96751 hypothetical protein F28P22.17
[imported] - Arabidopsis thaliana, partial (33%)
Length = 980
Score = 52.4 bits (124), Expect = 2e-07
Identities = 54/216 (25%), Positives = 82/216 (37%), Gaps = 1/216 (0%)
Frame = +1
Query: 61 DSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEE-SKQTIGASDDVYVGDIRD 119
D R VLVT G++V L S+ + LV+ + + + G + GD D
Sbjct: 289 DEVRDAVLVTDGDSEIGQMVILSLIVNKSRI--KALVKDKRVALEAFGNYVESMTGDTSD 462
Query: 120 TGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDA 179
+ A++G+ +I GF + G
Sbjct: 463 NRFLRKALRGVCTIICPNE-------GFISSVGS-------------------------- 543
Query: 180 AKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQD 239
GVK +VL+ + + S+ N + E L SGIPYTIIR G LQD
Sbjct: 544 --LQGVKHVVLLSQLSVYSGKTGIESMMKNNAKKLAEQDESVLRSSGIPYTIIRTGELQD 717
Query: 240 KEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQA 275
GG + K T +I++ DVA VC+++
Sbjct: 718 TPGGKQGFTFDKG----VAATGSISKEDVAFVCVKS 813
>TC93342 homologue to GP|20339364|gb|AAM19355.1 UOS1 {Pisum sativum},
partial (33%)
Length = 857
Score = 48.9 bits (115), Expect = 2e-06
Identities = 30/80 (37%), Positives = 44/80 (54%)
Frame = +3
Query: 211 ILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRELIIGKDDEILKTETRTIARPDVAE 270
IL +K K E + +SGIPY I+R L ++ G +LI + D I T I+R +VA
Sbjct: 270 ILTYKLKGEDLIRESGIPYVIVRPCALTEEPAGA-DLIFDQGDNI----TGKISREEVAR 434
Query: 271 VCIQALNFEEAQFKAFDLAS 290
+C+ AL A K F++ S
Sbjct: 435 MCVAALESPYACDKTFEVKS 494
>TC77041 similar to GP|3716262|emb|CAA03625.1 unnamed protein product
{unidentified}, partial (91%)
Length = 1447
Score = 40.8 bits (94), Expect = 7e-04
Identities = 50/174 (28%), Positives = 70/174 (39%), Gaps = 10/174 (5%)
Frame = +2
Query: 32 TSLQFSHSSSLSLATHKRVRT--RTSVVAMADSSRSTVLVTGAGGRTGKIVYKKLQERSS 89
+S+ FS + L HK + TS V+ D TV VTGAGG + K L ER
Sbjct: 104 SSVNFSTNIFFFLLKHKNMPAYDNTSSVSGGDQ---TVCVTGAGGFIASWLVKLLLERG- 271
Query: 90 QYIARGLVRTEES------KQTIGASD--DVYVGDIRDTGSIAPAIQGIDALIILTSAVP 141
Y RG VR E K+ GA + ++ D+ D SI + G + S V
Sbjct: 272 -YTVRGTVRNPEDPKNGHLKELEGARERLTLHKVDLLDLQSIQSVVHGCHGVFHTASPVT 448
Query: 142 LMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMG 195
+ P+ E G KN I A+ A V+++V S+G
Sbjct: 449 -----------DNPDEMLEPAVN-------GTKNVIIASAEAKVRRVVFTSSIG 556
>TC76963 GP|3716262|emb|CAA03625.1 unnamed protein product {unidentified},
complete
Length = 1468
Score = 37.7 bits (86), Expect = 0.006
Identities = 48/177 (27%), Positives = 69/177 (38%), Gaps = 14/177 (7%)
Frame = +1
Query: 33 SLQFSHSSS---LSLATHKRVRTRTSVVAMADSSR---STVLVTGAGGRTGKIVYKKLQE 86
S F H +S SL K + A A+SS T+ VTGAGG + K L E
Sbjct: 76 SHSFIHLTSPHLTSLYKKKEYMPAATAAAAAESSSVSGETICVTGAGGFIASWMVKLLLE 255
Query: 87 RSSQYIARGLVRTEES------KQTIGASDDVYV--GDIRDTGSIAPAIQGIDALIILTS 138
+ Y RG +R + K+ GA + + + D+ D S+ A+ G + S
Sbjct: 256 KG--YTVRGTLRNPDDPKNGHLKKLEGAKERLTLVKVDLLDLNSVKEAVNGCHGVFHTAS 429
Query: 139 AVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMG 195
V + PE E G KN I A A V+++V S+G
Sbjct: 430 PVT-----------DNPEEMVEPAVN-------GAKNVIIAGAEAKVRRVVFTSSIG 546
>BF636843 homologue to PIR|T10683|T106 hypothetical protein F3L17.100 -
Arabidopsis thaliana, partial (5%)
Length = 426
Score = 36.2 bits (82), Expect = 0.017
Identities = 20/66 (30%), Positives = 39/66 (58%), Gaps = 1/66 (1%)
Frame = +1
Query: 242 GGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLAS-KPEGAGTPTR 300
G R ++IG+ D+++ +R + VAE C+QAL+ E + + +++ S + EG G +
Sbjct: 52 GNDRAVLIGQGDKLVGEASRIV----VAEACVQALDLEATENQIYEVNSVEGEGPGNDAQ 219
Query: 301 DFKALF 306
++ LF
Sbjct: 220 KWQELF 237
>TC86516 homologue to GP|18087583|gb|AAL58922.1 AT4g35250/F23E12_190
{Arabidopsis thaliana}, partial (83%)
Length = 1499
Score = 34.3 bits (77), Expect = 0.064
Identities = 53/233 (22%), Positives = 92/233 (38%), Gaps = 8/233 (3%)
Frame = +2
Query: 65 STVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTI----GASDDVYVGDIRDT 120
+++LV GA G G+ + ++ + Y R LVR + GA+ V D+
Sbjct: 326 TSILVVGATGTLGRQIVRRALDEG--YDVRCLVRPRPAPADFLRDWGAT--VVNADLSKP 493
Query: 121 GSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAA 180
+I + G+ +I + P E P + VDW G+ I A
Sbjct: 494 ETIPATLVGVHTVIDCATGRP-----------EEPI---------KTVDWEGKVALIQCA 613
Query: 181 KAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDK 240
KA G+++ V S+ D + + ++ K E +L DSG+ + +IR G
Sbjct: 614 KAMGIQKYVFY-SIHNCDKHPEV------PLMEIKYCTENFLRDSGLNHIVIRLCGFMQG 772
Query: 241 EGGVRELIIGKDDEILKTETRT----IARPDVAEVCIQALNFEEAQFKAFDLA 289
G + I ++ + T+ T + D+A + AL E+ K A
Sbjct: 773 LIGQYAVPILEEKSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFA 931
>TC86434 similar to GP|2262100|gb|AAB63608.1| unknown protein {Arabidopsis
thaliana}, partial (83%)
Length = 1480
Score = 30.0 bits (66), Expect = 1.2
Identities = 22/79 (27%), Positives = 37/79 (45%), Gaps = 4/79 (5%)
Frame = -2
Query: 9 FVSATLSPNQCHKYCLPSSLKLLTSLQFSHSSSLSL----ATHKRVRTRTSVVAMADSSR 64
++ AT P +C PSS+KL+ L F HSS S+ + + ++ ++ S
Sbjct: 633 YLEAT*PPKEC-----PSSMKLVRFLAFLHSSKESMNHASVSSVLLHVKSGLLDPPQPS* 469
Query: 65 STVLVTGAGGRTGKIVYKK 83
ST ++ R K+ KK
Sbjct: 468 STAMILKREARASKLRKKK 412
>BM815228 similar to GP|22136000|gb unknown protein {Arabidopsis thaliana},
partial (43%)
Length = 705
Score = 29.6 bits (65), Expect = 1.6
Identities = 19/63 (30%), Positives = 30/63 (47%), Gaps = 2/63 (3%)
Frame = -1
Query: 17 NQCHKYCLPSSLKLLTSLQFSHSSSLSLATHKRVRTRTSVVAMADSSRST--VLVTGAGG 74
N +K+C PS + + S SSS ++A + + + M SS VL+T + G
Sbjct: 258 NLGYKHCFPSDISFQMTSSSSESSSFNIAATRLSYIKAASKRMPKSSSGVQLVLLTTSAG 79
Query: 75 RTG 77
R G
Sbjct: 78 RFG 70
>TC76636 homologue to GP|6625788|gb|AAF19398.1| dynamin homolog {Astragalus
sinicus}, partial (69%)
Length = 2420
Score = 29.3 bits (64), Expect = 2.0
Identities = 31/145 (21%), Positives = 60/145 (41%), Gaps = 1/145 (0%)
Frame = +3
Query: 74 GRTGKIVYKKLQERSSQYIARGLVRTEESK-QTIGASDDVYVGDIRDTGSIAPAIQGIDA 132
G+TGK+ Y K QE RG++ EE + + D +D S P +
Sbjct: 1059 GKTGKLGYTKKQEDRH---FRGVITLEECNIEEVPDESDPPPKSSKDKKSNGPDSSKVSL 1229
Query: 133 LIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVG 192
+ +TS VP + + + ++ +WI + + + AK ++ +
Sbjct: 1230 VFKITSRVP-----YKTVLKAHSAVVLKAESATDKTEWISKISSVIQAKGGQIR----LS 1382
Query: 193 SMGGTDLNNPLNSLGNGNILVWKRK 217
S GG+ + +SL +G++ R+
Sbjct: 1383 SEGGSAMR---HSLSDGSLDTMARR 1448
>TC87602 similar to GP|21741453|emb|CAD41397. OJ000223_09.9 {Oryza sativa},
partial (95%)
Length = 1263
Score = 29.3 bits (64), Expect = 2.0
Identities = 15/42 (35%), Positives = 26/42 (61%)
Frame = +2
Query: 165 PEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTDLNNPLNSL 206
P+QV I K +++ K GVK + +V +M G L+ P+++L
Sbjct: 692 PQQVSLIDVKKEVNFCKKVGVKVLGVVENMSG--LSQPISNL 811
>TC76539 aquaporin [Medicago truncatula]
Length = 1423
Score = 29.3 bits (64), Expect = 2.0
Identities = 22/73 (30%), Positives = 35/73 (47%), Gaps = 2/73 (2%)
Frame = +1
Query: 123 IAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYF-EDGAYPEQ-VDWIGQKNQIDAA 180
+AP G ++ + +P+ G NP + P F + A+ +Q + W+G I AA
Sbjct: 766 LAPLPIGFAVFMVHLATIPVTGTGINPARSFGPAVIFNNEKAWDDQWIYWVGP--FIGAA 939
Query: 181 KAAGVKQIVLVGS 193
AA Q +L GS
Sbjct: 940 VAAIYHQYILRGS 978
>CB893172 homologue to PIR|T45642|T45 FtsH metalloproteinase-like protein -
Arabidopsis thaliana, partial (25%)
Length = 661
Score = 29.3 bits (64), Expect = 2.0
Identities = 23/93 (24%), Positives = 44/93 (46%), Gaps = 2/93 (2%)
Frame = +3
Query: 52 TRTSVVAMADSSRSTVL--VTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGAS 109
+ + V+ +A ++R+ VL GR +IV + +R + +++ SK+ + +
Sbjct: 393 SNSPVIVLAATNRADVLDPALRRPGRFDRIVMVETPDRIGR---ESILKVHVSKKELPLA 563
Query: 110 DDVYVGDIRDTGSIAPAIQGIDALIILTSAVPL 142
DVY+GDI S+ G D ++ A L
Sbjct: 564 KDVYIGDI---ASMTTGFTGADLANLVNEAALL 653
>BF635925 similar to GP|8489806|gb chloroplast protein import component
Toc159 {Pisum sativum}, partial (6%)
Length = 389
Score = 28.9 bits (63), Expect = 2.7
Identities = 20/59 (33%), Positives = 28/59 (46%)
Frame = -1
Query: 12 ATLSPNQCHKYCLPSSLKLLTSLQFSHSSSLSLATHKRVRTRTSVVAMADSSRSTVLVT 70
A+ SP+ LPSS S S SSS +++T T T+ + SS +T L T
Sbjct: 335 ASASPSTALSTWLPSSTSTTRSTLLSASSSTAVSTWLSSSTSTTRSTLLSSSSATTLST 159
>TC90777 weakly similar to GP|15810655|gb|AAL07252.1 putative ubiquitin
carboxyl terminal hydrolase {Arabidopsis thaliana},
partial (21%)
Length = 962
Score = 28.5 bits (62), Expect = 3.5
Identities = 17/37 (45%), Positives = 19/37 (50%)
Frame = -3
Query: 9 FVSATLSPNQCHKYCLPSSLKLLTSLQFSHSSSLSLA 45
F SPN H + L SL L SL S S SLSL+
Sbjct: 123 FFLFNFSPNHNHNHTLSLSLSLSLSLSLSLSLSLSLS 13
>BG454712 similar to GP|6469135|emb| putative water channel protein {Cicer
arietinum}, partial (32%)
Length = 632
Score = 28.5 bits (62), Expect = 3.5
Identities = 19/67 (28%), Positives = 29/67 (42%), Gaps = 2/67 (2%)
Frame = -1
Query: 129 GIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAY--PEQVDWIGQKNQIDAAKAAGVK 186
G ++ +P+ PG NP + + DG + + W+G I AA AA
Sbjct: 620 GFAVFMVHLXTIPVTGPGINPARSFGSAVXYNDGKIWDDQWIFWVGP--FIGAAVAAIYH 447
Query: 187 QIVLVGS 193
Q +L GS
Sbjct: 446 QYILRGS 426
>BG583971 homologue to GP|6469135|emb putative water channel protein {Cicer
arietinum}, partial (42%)
Length = 808
Score = 28.1 bits (61), Expect = 4.6
Identities = 20/73 (27%), Positives = 32/73 (43%), Gaps = 2/73 (2%)
Frame = +2
Query: 123 IAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAY--PEQVDWIGQKNQIDAA 180
+AP G ++ + +P+ G NP + + DG + + W+G I AA
Sbjct: 53 LAPLPIGFAVFMVHLATIPVTGTGINPARSFGSAVIYNDGKIWDDQWIFWVGP--FIGAA 226
Query: 181 KAAGVKQIVLVGS 193
AA Q +L GS
Sbjct: 227 VAAIYHQYILRGS 265
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.134 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,689,114
Number of Sequences: 36976
Number of extensions: 90116
Number of successful extensions: 596
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 592
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 594
length of query: 313
length of database: 9,014,727
effective HSP length: 96
effective length of query: 217
effective length of database: 5,465,031
effective search space: 1185911727
effective search space used: 1185911727
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0147b.4


