

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0147a.4
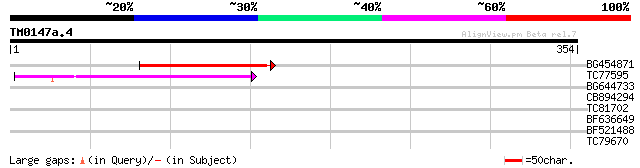
(354 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 89 3e-18
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 72 4e-13
BG644733 weakly similar to GP|15289942|db putative polyprotein {... 33 0.13
CB894294 similar to GP|20160527|dbj putative sexual differentiat... 30 1.8
TC81702 weakly similar to SP|P52369|VP40_HSVE2 Capsid protein P4... 29 2.4
BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativ... 29 3.1
BF521488 weakly similar to GP|18447433|gb RE28280p {Drosophila m... 28 5.4
TC79670 similar to PIR|A84635|A84635 probable calmodulin-binding... 27 9.2
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 88.6 bits (218), Expect = 3e-18
Identities = 44/85 (51%), Positives = 58/85 (67%)
Frame = +2
Query: 82 FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRRLIWTCPKNWSNCLAWAEFWF 141
F + L + GT L SSAYHP +DGQ+E +N+ E YLR L++T P WS WAE+W+
Sbjct: 38 FWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEYWY 217
Query: 142 NTNYNTSRNMSPFNALSGQDPLSLL 166
NT+YN S M+PF AL G+D LS+L
Sbjct: 218 NTSYNISAAMTPFKALYGRD-LSML 289
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial
(14%)
Length = 1708
Score = 71.6 bits (174), Expect = 4e-13
Identities = 49/154 (31%), Positives = 74/154 (47%), Gaps = 3/154 (1%)
Frame = +2
Query: 4 PTTVWENIYMDSIGGLPKSKGK--ETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEI 61
P + ++ MD I LP ++G+ + + V+VDRL+K L A A F+
Sbjct: 374 PNRLHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSKSVT-LEEMDTMEAEACAQRFLSCH 550
Query: 62 IRLHGFPSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLR 120
R HG P +IVSD S + F + G S++YHPQTDG TE N+ ++A LR
Sbjct: 551 YRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGTERWNQEIQAVLR 730
Query: 121 RLIWTCPKNWSNCLAWAEFWFNTNYNTSRNMSPF 154
+ NW + L + +N+S +PF
Sbjct: 731 AYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPF 832
>BG644733 weakly similar to GP|15289942|db putative polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (1%)
Length = 174
Score = 33.5 bits (75), Expect = 0.13
Identities = 19/43 (44%), Positives = 25/43 (57%)
Frame = -3
Query: 23 KGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIRLH 65
K E+I VVVDRLTK F+P +SA A + EI+ +H
Sbjct: 130 KSYESI*VVVDRLTKSTLFIPFKTSYSAK*YARILLDEIVCIH 2
>CB894294 similar to GP|20160527|dbj putative sexual differentiation process
protein Isp4 {Oryza sativa (japonica cultivar-group)},
partial (8%)
Length = 190
Score = 29.6 bits (65), Expect = 1.8
Identities = 15/46 (32%), Positives = 26/46 (55%)
Frame = +2
Query: 268 FMFPLLRRWWELISRFSSCLKCCLRITSS*FSLQMFWTLGKQLRGI 313
F+F RRWW+ R+S L L + ++ ++++L +LRGI
Sbjct: 26 FVFRYRRRWWQ---RYSYVLSAALDTGVAFMTVLLYFSLSLELRGI 154
>TC81702 weakly similar to SP|P52369|VP40_HSVE2 Capsid protein P40
[Contains: Assemblin (Protease) (EC 3.4.21.97); Capsid
assembly protein]., partial (5%)
Length = 1262
Score = 29.3 bits (64), Expect = 2.4
Identities = 12/40 (30%), Positives = 20/40 (50%)
Frame = +3
Query: 130 WSNCLAWAEFWFNTNYNTSRNMSPFNALSGQDPLSLLKGL 169
W NC+A+ W NTN + ++ N ++P L G+
Sbjct: 765 WKNCIAFVNDWMNTNVKCGQKIAEKNM---ENPQKFLLGI 875
>BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativa}, partial
(4%)
Length = 653
Score = 28.9 bits (63), Expect = 3.1
Identities = 17/56 (30%), Positives = 30/56 (53%), Gaps = 2/56 (3%)
Frame = +1
Query: 104 TDGQTEVVNRCLEAYLRRLIWTCPKNW--SNCLAWAEFWFNTNYNTSRNMSPFNAL 157
+D Q ++N LE +L L +T + + L WAE +NTN++ + +PF +
Sbjct: 7 SDEQAGLLNHTLETHL--LYFTSEQQGV*NFFLTWAECLYNTNFHRTAGCTPFKVV 168
>BF521488 weakly similar to GP|18447433|gb RE28280p {Drosophila
melanogaster}, partial (84%)
Length = 470
Score = 28.1 bits (61), Expect = 5.4
Identities = 13/31 (41%), Positives = 14/31 (44%), Gaps = 3/31 (9%)
Frame = +1
Query: 263 RYTWDFMFPLL---RRWWELISRFSSCLKCC 290
R W F F R WW RFS C+ CC
Sbjct: 307 RRRWWFCFSFR*C*RCWWRFPWRFSCCICCC 399
>TC79670 similar to PIR|A84635|A84635 probable calmodulin-binding protein
[imported] - Arabidopsis thaliana, partial (58%)
Length = 1213
Score = 27.3 bits (59), Expect = 9.2
Identities = 11/30 (36%), Positives = 17/30 (56%)
Frame = -1
Query: 52 EVAAGFIQEIIRLHGFPSTIVSDHDSFPE* 81
++ F + LHGF S ++S+H FP *
Sbjct: 523 QLEVAFSPRVPLLHGFQSALLSNHHQFPH* 434
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.349 0.154 0.574
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,066,904
Number of Sequences: 36976
Number of extensions: 241109
Number of successful extensions: 2274
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 2251
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2273
length of query: 354
length of database: 9,014,727
effective HSP length: 97
effective length of query: 257
effective length of database: 5,428,055
effective search space: 1395010135
effective search space used: 1395010135
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0147a.4


