

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0137.11
(178 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
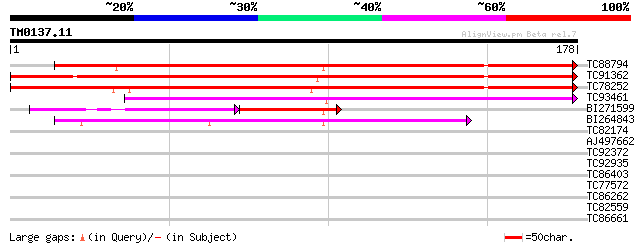
Score E
Sequences producing significant alignments: (bits) Value
TC88794 similar to GP|15524453|emb|CAC69349. unnamed protein pro... 177 2e-45
TC91362 weakly similar to GP|15524453|emb|CAC69349. unnamed prot... 175 7e-45
TC78252 similar to GP|15524450|emb|CAC69348. unnamed protein pro... 147 3e-36
TC93461 81 2e-16
BI271599 weakly similar to GP|22212790|gb| invertase inhibitor-l... 43 4e-08
BI264843 45 1e-05
TC82174 similar to GP|4097684|gb|AAD00154.1| ubiquitin conjugati... 39 0.001
AJ497662 38 0.002
TC92372 weakly similar to GP|9759184|dbj|BAB09799.1 pectinestera... 36 0.007
TC92935 weakly similar to GP|17933303|gb|AAL48234.1 At1g12650/T1... 29 0.86
TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF028... 28 1.9
TC77572 similar to GP|9558523|dbj|BAB03441.1 ESTs AU078742(C1188... 27 5.6
TC86262 homologue to GP|22335691|dbj|BAC10548. cullin-like prote... 26 7.3
TC82559 weakly similar to PIR|T48313|T48313 hypothetical protein... 26 7.3
TC86661 similar to GP|9798599|dbj|BAB11737.1 serine/threonine pr... 26 9.6
>TC88794 similar to GP|15524453|emb|CAC69349. unnamed protein product
{Glycine max}, partial (79%)
Length = 638
Score = 177 bits (449), Expect = 2e-45
Identities = 89/166 (53%), Positives = 118/166 (70%), Gaps = 2/166 (1%)
Frame = +1
Query: 15 IVVATISMSIGHCRVLQP-SDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIM 73
+ + +M HC ++ P S+A L+ +TCK TP + C+ +L++DPRSS ADVTGLALIM
Sbjct: 31 LAIILCTMVSSHCIIIHPKSNANLIQQTCKQTPNYANCIHYLKSDPRSSDADVTGLALIM 210
Query: 74 VDVIQAKTNGVLNKISQLLKGGGD-KPALNSCQGRYNAILKADIPQATQALKTGNPKFAE 132
VD+I++K N LNKI+QL+KG D K ALNSC GRY AIL AD+P++ ALK G+PKFAE
Sbjct: 211 VDIIKSKANTALNKINQLIKGSHDQKEALNSCAGRYRAILVADVPKSVAALKQGDPKFAE 390
Query: 133 DGVADAGVEANTCESGFSSGKSPLTSENNGMHIATEVARAIIRNLL 178
DG D +EA TCE+GF GKSP++ EN MH + AI++ LL
Sbjct: 391 DGANDVAIEATTCENGF-KGKSPISDENIDMHDVAVITAAIVKQLL 525
>TC91362 weakly similar to GP|15524453|emb|CAC69349. unnamed protein product
{Glycine max}, partial (64%)
Length = 611
Score = 175 bits (444), Expect = 7e-45
Identities = 89/180 (49%), Positives = 122/180 (67%), Gaps = 2/180 (1%)
Frame = +1
Query: 1 MENLKSLSLICNIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPR 60
++N+ F+++ T + + R +QP+DA L+ +TCKNTP + C+Q+L +DP+
Sbjct: 7 LKNMTMFKPFALFFLILCTF-LVVTQSRTIQPNDANLIQQTCKNTPNYALCIQYLNSDPK 183
Query: 61 SSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGG--GDKPALNSCQGRYNAILKADIPQ 118
+ SAD+ GL+LIMV+VI+ K LNKI QLLK K AL SC+ +YN I+ AD+ +
Sbjct: 184 APSADIRGLSLIMVNVIKTKATATLNKIRQLLKTSPPSQKGALTSCESKYNTIIVADVAE 363
Query: 119 ATQALKTGNPKFAEDGVADAGVEANTCESGFSSGKSPLTSENNGMHIATEVARAIIRNLL 178
AT+AL+ GNPKFAEDG +DA VE CE GF SG SPLT++NN MH +ARAII NLL
Sbjct: 364 ATEALQKGNPKFAEDGASDADVEQEACEGGF-SGNSPLTADNNVMHNVATIARAIIPNLL 540
>TC78252 similar to GP|15524450|emb|CAC69348. unnamed protein product
{Glycine max}, partial (66%)
Length = 1395
Score = 147 bits (370), Expect = 3e-36
Identities = 90/183 (49%), Positives = 114/183 (62%), Gaps = 5/183 (2%)
Frame = +1
Query: 1 MENLKSLSLICNIFIVVATISMSIGHCRVLQ-PSDAQ---LVAKTCKNTPYPSACLQFLQ 56
M N K L+L + +V IS+ R PS + L+ TCK TP + C Q L+
Sbjct: 205 MTNFKPLTLCLFLQAIVIIISLPTIQSRSTNFPSKDEKLSLIENTCKKTPNYNVCFQSLK 384
Query: 57 ADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQLLK-GGGDKPALNSCQGRYNAILKAD 115
A +S+ DVTGLA IMV V++AK N LNKI QL + G G + AL+SC +Y AIL AD
Sbjct: 385 AYSGTSAGDVTGLAQIMVKVMKAKANDGLNKIHQLQRLGNGARKALSSCGDKYRAILIAD 564
Query: 116 IPQATQALKTGNPKFAEDGVADAGVEANTCESGFSSGKSPLTSENNGMHIATEVARAIIR 175
IPQA +AL+ G+PKFAEDG DA EA CES F +GKSPLT +NN MH + V +I+R
Sbjct: 565 IPQAIEALEKGDPKFAEDGANDAANEATYCESEF-NGKSPLTKQNNAMHDVSAVTSSIVR 741
Query: 176 NLL 178
LL
Sbjct: 742 QLL 750
>TC93461
Length = 612
Score = 81.3 bits (199), Expect = 2e-16
Identities = 51/145 (35%), Positives = 70/145 (48%), Gaps = 3/145 (2%)
Frame = +2
Query: 37 LVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGG 96
LV + CK TP+ C L A+P + +D G+ALIMV+ I A L+ I +L+K
Sbjct: 125 LVDQICKQTPFYDLCSSILHANPLAPKSDPKGMALIMVNDILANATDTLSYIEELIKQTT 304
Query: 97 DK---PALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCESGFSSGK 153
DK L C Y ++K +PQA A+ G FA + DA E C F
Sbjct: 305 DKDLEQQLAFCAESYIPVVKYILPQAADAISQGRYGFASYSIVDAEKEIGACNKKFPGSS 484
Query: 154 SPLTSENNGMHIATEVARAIIRNLL 178
S L N+ M +VA AI++ LL
Sbjct: 485 SLLGDRNSIMQKLVDVAAAIVKLLL 559
>BI271599 weakly similar to GP|22212790|gb| invertase inhibitor-like
protein {Ipomoea batatas}, partial (13%)
Length = 307
Score = 43.1 bits (100), Expect(2) = 4e-08
Identities = 25/66 (37%), Positives = 39/66 (58%)
Frame = +3
Query: 7 LSLICNIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADV 66
LS +F V A IS+ + +QP L+ +TCK TP + C+++L++DPR+S AD+
Sbjct: 21 LSSCALLFHVTAQISIKK---QTMQP----LIQQTCKQTPNYALCIKYLKSDPRTSDADL 179
Query: 67 TGLALI 72
A I
Sbjct: 180VNYAXI 197
Score = 30.0 bits (66), Expect(2) = 4e-08
Identities = 17/35 (48%), Positives = 22/35 (62%), Gaps = 3/35 (8%)
Frame = +1
Query: 73 MVDVIQAKTNGVLNKISQLLKGGGD---KPALNSC 104
MVD I+ T NK++QLLKGG + AL+SC
Sbjct: 199 MVDRIKTTTITAYNKVTQLLKGGHELYQTEALSSC 303
>BI264843
Length = 655
Score = 45.4 bits (106), Expect = 1e-05
Identities = 34/141 (24%), Positives = 65/141 (45%), Gaps = 10/141 (7%)
Frame = +2
Query: 15 IVVATIS--MSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRS-----SSADVT 67
I++ TIS I + QP L+ C+ TP+ C+ L+++ + S+ D++
Sbjct: 62 IILLTISSIFFIPPIQCHQPPKLDLIKAICRKTPHYHLCVITLKSNLYAVKYIASNTDIS 241
Query: 68 GLALIMVDVIQAKTNGVLNKISQLLKGGGD---KPALNSCQGRYNAILKADIPQATQALK 124
G + + V+ + +L+++ + D K AL++C Y I K +PQA + +
Sbjct: 242 GFVRVTLKVVATRAPVILHQLQTVQVQTKDIQLKAALDNCIASYTKISKELVPQAQKCVD 421
Query: 125 TGNPKFAEDGVADAGVEANTC 145
+ + AG A+TC
Sbjct: 422 KSDYNGVKQSATTAGNLADTC 484
>TC82174 similar to GP|4097684|gb|AAD00154.1| ubiquitin conjugating enzyme
{Metarhizium anisopliae}, partial (17%)
Length = 1031
Score = 38.5 bits (88), Expect = 0.001
Identities = 31/149 (20%), Positives = 61/149 (40%), Gaps = 11/149 (7%)
Frame = +1
Query: 7 LSLICNIFIVVATISMSI-----GHCR------VLQPSDAQLVAKTCKNTPYPSACLQFL 55
LSL+ + I+ +TIS ++ H L+ + Q ++ TC T +PS C+ +L
Sbjct: 481 LSLLAVLLIIASTISAAMLTGIHSHTTSEPKNPTLRRNPTQAISNTCSKTRFPSLCINYL 660
Query: 56 QADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGDKPALNSCQGRYNAILKAD 115
P S+ A L I +++ + L + + G P + + +L
Sbjct: 661 LDFPDSTGASEKDLVHISLNMTLQHLSKALYTSASISSTVGINPYIRAAYTDCLELLDNS 840
Query: 116 IPQATQALKTGNPKFAEDGVADAGVEANT 144
+ +AL + P + +G ++T
Sbjct: 841 VDALARALTSAVPSSSSNGAVKPLTSSST 927
>AJ497662
Length = 629
Score = 37.7 bits (86), Expect = 0.002
Identities = 30/143 (20%), Positives = 60/143 (40%), Gaps = 6/143 (4%)
Frame = +2
Query: 38 VAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQL-LKGGG 96
V + CK PS C L++ P + D+ LA +DV++ + ++ I++L + G
Sbjct: 83 VQEICKKATNPSFCSTILESKPGGAGKDLVSLASYTIDVLRTNVSNTISLITKLAAQSGS 262
Query: 97 DKPALN---SCQGRYNAILKA--DIPQATQALKTGNPKFAEDGVADAGVEANTCESGFSS 151
D N +C R+ A ++ +A + LK + + + C S SS
Sbjct: 263 DSLKQNRYKNCLSRFGMDDGALGEVEEAQKVLKDSDYNGVNMHITSVMTAVDECLSKDSS 442
Query: 152 GKSPLTSENNGMHIATEVARAII 174
+ + + ++A+ I+
Sbjct: 443 PDNDTSLLPKDVECVNQIAQIIL 511
>TC92372 weakly similar to GP|9759184|dbj|BAB09799.1 pectinesterase
{Arabidopsis thaliana}, partial (10%)
Length = 605
Score = 36.2 bits (82), Expect = 0.007
Identities = 20/73 (27%), Positives = 35/73 (47%), Gaps = 10/73 (13%)
Frame = +3
Query: 2 ENLKSLSLICNIFIVVATISMSI----------GHCRVLQPSDAQLVAKTCKNTPYPSAC 51
+ L LS++ + I+ + IS ++ + +L Q +++TC T YPS C
Sbjct: 297 KKLIMLSILAAVLIIASAISAALITVVRSRASSNNSNLLHSKPTQAISRTCSKTRYPSLC 476
Query: 52 LQFLQADPRSSSA 64
+ L P S+SA
Sbjct: 477 INSLLDFPGSTSA 515
>TC92935 weakly similar to GP|17933303|gb|AAL48234.1 At1g12650/T12C24_1
{Arabidopsis thaliana}, partial (45%)
Length = 725
Score = 29.3 bits (64), Expect = 0.86
Identities = 20/58 (34%), Positives = 27/58 (46%)
Frame = +2
Query: 75 DVIQAKTNGVLNKISQLLKGGGDKPALNSCQGRYNAILKADIPQATQALKTGNPKFAE 132
DVIQA GV + + L G D RYN + + D+P QALK K+ +
Sbjct: 368 DVIQAPKKGVRDPRFESLCGTFDNDGFRK---RYNFLYENDLPAERQALKRQLRKYKD 532
>TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1929
Score = 28.1 bits (61), Expect = 1.9
Identities = 22/89 (24%), Positives = 45/89 (49%), Gaps = 6/89 (6%)
Frame = +3
Query: 8 SLICN---IFIVVATISMSIGHCRVLQPSDAQLVAK--TCKNTPYPSACLQFLQADPRSS 62
S +CN IF+++ ++S++ V P+ ++ CK+TP P+ C L P+++
Sbjct: 21 SKLCNPLSIFLILLSVSIAFSQNTVNNPNPITPISPGAACKSTPDPTYCKSIL--PPQNA 194
Query: 63 SADVTGLALIMVDVIQA-KTNGVLNKISQ 90
+ G + + Q+ K + ++NK Q
Sbjct: 195 NVYDYGRFSVKKSLSQSRKFSDLINKYLQ 281
>TC77572 similar to GP|9558523|dbj|BAB03441.1 ESTs AU078742(C11888)
C26225(C11888) correspond to a region of the predicted
gene.~Similar to Oryza, partial (5%)
Length = 2535
Score = 26.6 bits (57), Expect = 5.6
Identities = 24/74 (32%), Positives = 33/74 (44%), Gaps = 8/74 (10%)
Frame = +1
Query: 82 NGVLNKISQLLKGGGDKPALNSCQGRYNAILKADIPQATQALKTGN-----PKFAEDGVA 136
N VLNKISQ+L+ K N R ++ D+ Q +K N P+ + +
Sbjct: 85 NDVLNKISQILQ----KAQKNQSSRRSLRLILKDLSPVVQDIKQYNDHLDHPREEINSLI 252
Query: 137 ---DAGVEANTCES 147
DAG A TC S
Sbjct: 253 EENDAGESACTCSS 294
>TC86262 homologue to GP|22335691|dbj|BAC10548. cullin-like protein1 {Pisum
sativum}, complete
Length = 2332
Score = 26.2 bits (56), Expect = 7.3
Identities = 27/91 (29%), Positives = 39/91 (42%), Gaps = 3/91 (3%)
Frame = +2
Query: 23 SIGHCRV---LQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQA 79
S+G C + P +LV T + SA L F +D S S +T L L+ DVI+
Sbjct: 1793 SLGTCNISGKFDPKTVELVVTTYQ----ASALLLFNSSDRLSYSEIMTQLNLLDEDVIRL 1960
Query: 80 KTNGVLNKISQLLKGGGDKPALNSCQGRYNA 110
+ K L+K K L + +NA
Sbjct: 1961 LHSLSCAKYKILIKEPNTKTILPTDYFEFNA 2053
>TC82559 weakly similar to PIR|T48313|T48313 hypothetical protein F9G14.230
- Arabidopsis thaliana, partial (10%)
Length = 693
Score = 26.2 bits (56), Expect = 7.3
Identities = 13/26 (50%), Positives = 14/26 (53%)
Frame = -3
Query: 85 LNKISQLLKGGGDKPALNSCQGRYNA 110
LNK S LL G PA N C + NA
Sbjct: 433 LNKFSLLLPSSGPIPAANFCS*KINA 356
>TC86661 similar to GP|9798599|dbj|BAB11737.1 serine/threonine protein kinase
{Arabidopsis thaliana}, partial (58%)
Length = 1840
Score = 25.8 bits (55), Expect = 9.6
Identities = 15/36 (41%), Positives = 20/36 (54%)
Frame = -3
Query: 31 QPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADV 66
QPS +L+ K+ +P P + LQF P SSS V
Sbjct: 1418 QPSQ-RLIGKSQSQSPNPHSALQFPPHIPSSSSPQV 1314
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,025,076
Number of Sequences: 36976
Number of extensions: 59022
Number of successful extensions: 294
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 287
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 289
length of query: 178
length of database: 9,014,727
effective HSP length: 90
effective length of query: 88
effective length of database: 5,686,887
effective search space: 500446056
effective search space used: 500446056
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0137.11


