

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0137.1
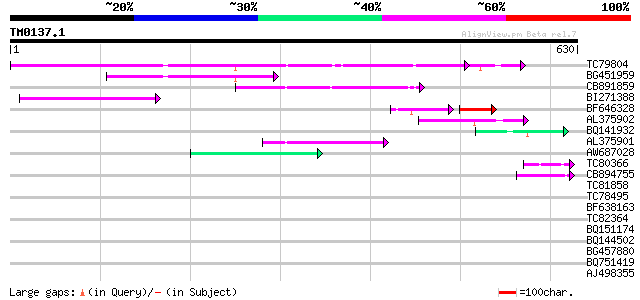
(630 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like... 240 2e-68
BG451959 homologue to GP|6175165|gb| Mutator-like transposase {A... 114 1e-25
CB891859 similar to GP|6175165|gb| Mutator-like transposase {Ara... 92 8e-19
BI271388 similar to GP|10177186|db mutator-like transposase-like... 86 6e-17
BF646328 similar to PIR|F84533|F845 Mutator-like transposase [im... 55 5e-15
AL375902 similar to GP|9759134|dbj mutator-like transposase-like... 79 5e-15
BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 47 3e-05
AL375901 similar to GP|9759134|dbj mutator-like transposase-like... 46 5e-05
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 43 4e-04
TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like... 42 5e-04
CB894755 similar to GP|9757890|db phosphoribosylanthranilate tra... 42 7e-04
TC81858 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 41 0.001
TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 ... 40 0.003
BF638163 39 0.008
TC82364 similar to GP|20268740|gb|AAM14073.1 unknown protein {Ar... 39 0.008
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 38 0.010
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 37 0.023
BG457880 weakly similar to GP|1063689|gb|A ORF 66; this sequence... 37 0.023
BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania maj... 37 0.023
AJ498355 37 0.030
>TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like
transposase {Arabidopsis thaliana}, partial (57%)
Length = 2129
Score = 240 bits (612), Expect(2) = 2e-68
Identities = 159/518 (30%), Positives = 255/518 (48%), Gaps = 9/518 (1%)
Frame = +1
Query: 2 EFKFKLGMEFTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAICRRR-CGFLIFLSKVG 60
+ F +G EF ++ FR A+++ ++ EL+ +K+D +R A C C + I K+
Sbjct: 166 DHNFVVGQEFPDVKAFRNAIKEAAIAQHFELRIIKSDLIRYFAKCASEGCPWRIRAVKLP 345
Query: 61 GKHTYSIKTLIPEHTCSR--VYNNRSATSDFVASKLKEKVSIVRKYRLSEVVDEMRIHYS 118
T++I++L HTC R + + A+ D++ S ++E++ Y+ +++ ++ Y
Sbjct: 346 NASTFTIRSLEGTHTCGRNALNGHHQASVDWIVSFIEERLRDNINYKPKDILHDIHKQYG 525
Query: 119 TGITTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQISQP 178
I +AWRA+++ LA + G + + L F E+K+ +PG+ K +
Sbjct: 526 ITIPYKQAWRAKERGLAAIYGSSEEGFYLLPSFCEEIKKTNPGSVAKVFTTG-----ADS 690
Query: 179 RFNRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVE 238
RF R +I GFV GC P + L G LK++Y L A D + FP+AFAVV+
Sbjct: 691 RFQRLFISFYASIHGFVNGCLPIVALGGIQLKSKYLSTFLSATSFDADGGLFPLAFAVVD 870
Query: 239 SECKESWKWFL----ELLLADIGGLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYA 294
E ESW WFL L + + + IF+SD QKG++ P H FC+RHL
Sbjct: 871 VENDESWTWFLSELHNALEVNTECMPQIIFLSDGQKGIVDAIRRKFPRSSHAFCMRHLSE 1050
Query: 295 NFKKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAF- 353
N K+F +I L+ AA AT ++ F EKM E++EV+ A WL W F
Sbjct: 1051NIGKEFKNSRLIH-LLWSAAYATTINAFREKMAEIEEVSPNASMWLQHFHPSQWALVYFE 1227
Query: 354 -TLYPKCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFM 412
T Y L +N+ E FN IL A++ PI+ +I+ I+S L F K +
Sbjct: 1228GTRYGH---LSSNIEE-FNKWILEAQELPIIQVIERIQSKLKTEFDDRRLKSSSWCSVLT 1395
Query: 413 PKPLKRLAWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGIPCR 472
P +R+ ++ + ++EV S D IV++ SC+C W L GIPC
Sbjct: 1396PSSERRMVEAINRASTYQVLKSDEVEFEVISADRSD--IVNIGSHSCSCRDWQLYGIPCS 1569
Query: 473 HAVAAISFNGGHAESYIDSYYGRDAYQATYAHGITPIN 510
HAVAA+ + +Y + +Y+ T G+TP++
Sbjct: 1570HAVAALISSRKDVYAYTAKCFTVASYRDTVCRGVTPLS 1683
Score = 38.5 bits (88), Expect(2) = 2e-68
Identities = 23/73 (31%), Positives = 31/73 (41%), Gaps = 6/73 (8%)
Frame = +3
Query: 507 TPINGKRMWPYTPDA------PILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRC 560
TP+ GK W A + PP ++R PGRP+K R ++ +R C
Sbjct: 1674 TPVPGKLEWRTDESALDNDIAVVRPPKFRRPPGRPEKKR------ICVEDHNRDKHTVHC 1835
Query: 561 GRCGTIGHNIRTC 573
RC GH TC
Sbjct: 1836 SRCNQTGHYKTTC 1874
>BG451959 homologue to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (28%)
Length = 658
Score = 114 bits (285), Expect = 1e-25
Identities = 63/195 (32%), Positives = 103/195 (52%), Gaps = 4/195 (2%)
Frame = +2
Query: 108 EVVDEMRIHYSTGITTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFH 167
E+++E+ + ++ +AWR ++ +A + G Y L ++ A +KR +PG+ +
Sbjct: 86 EILEEIHRVHGITLSYKQAWRGKEHIMAAMRGSFEEGYRLLPQYCAHVKRTNPGSIASVY 265
Query: 168 CLPPTAQISQPRFNRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPND 227
P S F R +I + G + CRP +GLD +LK++Y G LL A G D +
Sbjct: 266 GNP-----SDNCFQRLFISFQASIYGLLNACRPLLGLDRIYLKSKYLGTLLLATGFDGDG 430
Query: 228 QYFPIAFAVVESECKESWKWFL----ELLLADIGGLRRWIFISDQQKGLLSVFDELMPGV 283
FP+AF VV+ E ++W WFL LL + + R +SD+Q+G++ + P
Sbjct: 431 ALFPLAFGVVDEENDDNWMWFLSKLHNLLEINTENMPRLTILSDRQQGIVDGVEANFPTA 610
Query: 284 EHRFCLRHLYANFKK 298
H FC+RHL NF+K
Sbjct: 611 FHGFCMRHLSDNFRK 655
>CB891859 similar to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (27%)
Length = 625
Score = 91.7 bits (226), Expect = 8e-19
Identities = 61/211 (28%), Positives = 108/211 (50%)
Frame = +1
Query: 251 LLLADIGGLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYANFKKKFGGGVVIRDLM 310
LL + + R +SD+Q+G++ + P H FC+RHL +F+K+F +++ +L+
Sbjct: 1 LLEVNTENMPRLTILSDRQQGIVDGVEANFPTAFHGFCMRHLSDSFRKEFNNTMLV-NLL 177
Query: 311 MQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTLYPKCDVLMNNLSEAF 370
+AA + FE K+ E++E++++A W+ P + W F + + L N+ EA
Sbjct: 178 WEAANCLTIIEFEGKVMEIEEISQDAAYWIRRVPPRLWATAYFEGH-RFGHLTANIVEAL 354
Query: 371 NATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPKPLKRLAWETKSSNNWV 430
N+ IL A PI+ M++ IR LM F E ++ +P + +A + + +
Sbjct: 355 NSWILEASGLPIIQMMECIRRQLMTWFNERRETSMQWTSILVPSAERSVAEALERARTYQ 534
Query: 431 PQACGHAKYEVKSVLDGDQFIVDLRQRSCTC 461
A++EV S +G IVD+R R C C
Sbjct: 535 VLRANEAEFEVIS-HEGTN-IVDIRNRCCLC 621
>BI271388 similar to GP|10177186|db mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (26%)
Length = 711
Score = 85.5 bits (210), Expect = 6e-17
Identities = 49/160 (30%), Positives = 81/160 (50%), Gaps = 3/160 (1%)
Frame = +2
Query: 11 FTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAICRRR-CGFLIFLSKVGGKHTYSIKT 69
F S +FR+AL +S+ +G ++ KND RV C+ + C + I+ SK+ IK
Sbjct: 149 FNSFSEFREALHKYSIAHGFAYRYKKNDSHRVTVKCKSQGCPWRIYASKLSTTQLICIKK 328
Query: 70 LIPEHTC--SRVYNNRSATSDFVASKLKEKVSIVRKYRLSEVVDEMRIHYSTGITTWRAW 127
+ +HTC S V AT +V + +KEK+ YR ++ D+++ Y + +AW
Sbjct: 329 MTRDHTCEGSAVKAGYRATRGWVGNIIKEKLKASPNYRPKDIADDIKREYGIQLNYSQAW 508
Query: 128 RARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFH 167
RA++ A ++G YT L F ++K +PG H
Sbjct: 509 RAKEIAREQLQGSYKEAYTQLPFFCEKIKETNPGEFCNIH 628
>BF646328 similar to PIR|F84533|F845 Mutator-like transposase [imported] -
Arabidopsis thaliana, partial (3%)
Length = 636
Score = 55.1 bits (131), Expect(2) = 5e-15
Identities = 27/73 (36%), Positives = 41/73 (55%), Gaps = 3/73 (4%)
Frame = -1
Query: 424 KSSNNWVPQACGHAKYEVKSVL---DGDQFIVDLRQRSCTCNWWDLVGIPCRHAVAAISF 480
KS W +A H E+ + + +++IV+L QR+C C W+L GIPC H + I
Sbjct: 492 KSCRGW--KATWHGDMEMNNFNVSNETNKYIVNLAQRTCACRKWNLTGIPCAHVIPCIWH 319
Query: 481 NGGHAESYIDSYY 493
NG E+++ SYY
Sbjct: 318 NGLADENFVSSYY 280
Score = 44.3 bits (103), Expect(2) = 5e-15
Identities = 19/42 (45%), Positives = 27/42 (64%)
Frame = -3
Query: 500 ATYAHGITPINGKRMWPYTPDAPILPPLYKRKPGRPKKLRRR 541
ATY+H + P +G ++WP T I P +R GRPKKLR++
Sbjct: 175 ATYSHIVLPSSGPKLWPVTNTEHINPLAKRRSAGRPKKLRKK 50
>AL375902 similar to GP|9759134|dbj mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (20%)
Length = 542
Score = 79.0 bits (193), Expect = 5e-15
Identities = 45/133 (33%), Positives = 60/133 (44%), Gaps = 11/133 (8%)
Frame = +2
Query: 455 RQRSCTCNWWDLVGIPCRHAVAAISFNGGHAESYIDSYYGRDAYQATYAHGITPINGKRM 514
R R C+C W L G PC HA AA+ G +A + + + +Y+ Y+ I PI K
Sbjct: 2 RSRECSCRRWQLYGYPCAHAAAALISCGHNAHMFAEPCFTVQSYRMAYSQMIYPIPDKSQ 181
Query: 515 W-----------PYTPDAPILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRC 563
W D I PP +R PGRPKK R + RP +CGRC
Sbjct: 182 WREHGEGAEGGGGARVDIVIHPPKIRRPPGRPKKKVLR------VENFKRPKRVVQCGRC 343
Query: 564 GTIGHNIRTCTNP 576
+GH+ + CT P
Sbjct: 344 HMLGHSQKKCTMP 382
>BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (30%)
Length = 1338
Score = 46.6 bits (109), Expect = 3e-05
Identities = 32/106 (30%), Positives = 39/106 (36%), Gaps = 3/106 (2%)
Frame = -1
Query: 518 TPDAPILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTC---T 574
+P P PP +PGRP+ P S C CG + T
Sbjct: 450 SPPPPPHPPTSPPRPGRPRSSTLAPPPHRQFHSPSA-----LCTHCGIPPPAVPPPPPPT 286
Query: 575 NPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPP 620
PP+S P P P PP QP+ + PP P PPQPP
Sbjct: 285 PPPSSPPALVSPPPRPFLFPPPPNQPSPSPSPPPPPPLPPPPPQPP 148
Score = 36.2 bits (82), Expect = 0.039
Identities = 31/112 (27%), Positives = 38/112 (33%), Gaps = 10/112 (8%)
Frame = -2
Query: 519 PDAPILPPLYKR---KPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTCTN 575
P P+ PP + R P RP LR P G RP+ R G +
Sbjct: 494 PPHPVRPPTHARLRSPPLRPPTLR--PPPPAPVAHGHRPSPPRRIGSSTPPPPFVPIAGF 321
Query: 576 PPTSQPTEDVP---ETEPTEAAQPPQQPTE----AAQPPQQPTEATQPPQPP 620
PP P P P ++ PP P+ PP P PP PP
Sbjct: 320 PPPPSPPPPPPLPLHPPPQLSSLPPHAPSSFLLPLTSPPPPPPPPHPPPSPP 165
Score = 32.0 bits (71), Expect = 0.74
Identities = 18/52 (34%), Positives = 21/52 (39%), Gaps = 7/52 (13%)
Frame = -1
Query: 575 NPPTSQ-------PTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQP 619
NPP SQ PT P P + PP + PP P T PP+P
Sbjct: 558 NPPASQSPATLNRPTSHTPACPPP-SRPPPHPRAPSLSPPPPPHPPTSPPRP 406
Score = 30.0 bits (66), Expect = 2.8
Identities = 15/58 (25%), Positives = 25/58 (42%), Gaps = 4/58 (6%)
Frame = -3
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQP----PQQPTEATQPPQPPTVAAQNPVV 629
PP++ P D+P + P+ P +A P P P + P +PP +P +
Sbjct: 448 PPSAPPPSDLPPPPRSPTVIDPRPPAASAVPLPLRPLYPLRDSPPRRPPPPPPHSPSI 275
Score = 29.6 bits (65), Expect = 3.7
Identities = 18/50 (36%), Positives = 20/50 (40%), Gaps = 3/50 (6%)
Frame = -2
Query: 574 TNPP-TSQPTEDVPETEPTEAA--QPPQQPTEAAQPPQQPTEATQPPQPP 620
T+PP T P P PT A PP +P PP P P PP
Sbjct: 524 TDPPATPLPAPPHPVRPPTHARLRSPPLRPPTLRPPPPAPVAHGHRPSPP 375
Score = 28.9 bits (63), Expect = 6.2
Identities = 16/51 (31%), Positives = 22/51 (42%)
Frame = -3
Query: 577 PTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
PT QP +P P +A PP + P++ PP+ PTV P
Sbjct: 523 PTHQPHPCLPP--PIPSAPPPTRAFALPPSAPPPSDLPPPPRSPTVIDPRP 377
>AL375901 similar to GP|9759134|dbj mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (27%)
Length = 493
Score = 45.8 bits (107), Expect = 5e-05
Identities = 32/140 (22%), Positives = 59/140 (41%)
Frame = +3
Query: 281 PGVEHRFCLRHLYANFKKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWL 340
P H FCLR++ NF+ F ++ ++ A A FE K+ E+ EV+++ W
Sbjct: 12 PSASHGFCLRYVSENFRDTFKNTKLV-NIFWNAVYALTAAEFESKITEMIEVSQDVISWF 188
Query: 341 MTHPKKSWCKHAFTLYPKCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAAL 400
P W A+ + ++E L + P++ M+++IR + F
Sbjct: 189 QHFPPFLWAV-AYFDGVRYGHFTLGVTELLYNWALECHELPVVQMMEYIRQQMTSWFNDR 365
Query: 401 NEKFQRYNHPFMPKPLKRLA 420
E + +P KR++
Sbjct: 366 REVGMEWTSILVPSAEKRIS 425
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 42.7 bits (99), Expect = 4e-04
Identities = 34/148 (22%), Positives = 58/148 (38%), Gaps = 2/148 (1%)
Frame = +1
Query: 202 IGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVESECKESWKWFLELLLADIGGLRR 261
+ D + K +Y L G + + Q A++E E ES+KW L L +
Sbjct: 7 LAFDTTYKKNKYNYPLCIFSGCNHHSQTIIFGVALLEDETIESYKWVLNRFLECMENKFP 186
Query: 262 WIFISDQQKGLLSVFDELMPGVEHRFCLRHLYANFKKKFGGGVVIRDLMMQAAKATYVHL 321
++D + ++ P HR C HL+ N ++ +
Sbjct: 187 KAVVTDGDGSMREAIKQVFPDASHRLCAWHLHKNAQENIKKTPFLEGFRKAMYSNFTPEQ 366
Query: 322 FEEKMRELKEVNE-EAHDWLM-THPKKS 347
FE+ EL + NE E + W++ T+ KS
Sbjct: 367 FEDFWSELIQKNELEGNAWVIKTYANKS 450
>TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like protein
{Arabidopsis thaliana}, partial (16%)
Length = 814
Score = 42.4 bits (98), Expect = 5e-04
Identities = 22/56 (39%), Positives = 28/56 (49%)
Frame = +3
Query: 572 TCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
T NPP P+ P T P + PP T +A PP P+ + PP PPT A +P
Sbjct: 45 TPANPPPVTPSAPPPST-PATPSSPPPSTTPSAPPPSTPSNS--PPSPPTTPAISP 203
Score = 36.6 bits (83), Expect = 0.030
Identities = 18/56 (32%), Positives = 26/56 (46%)
Frame = +3
Query: 572 TCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
T ++PP S P P T +A PP P + PP T + PP P+ + +P
Sbjct: 21 TPSSPPPSTPANPPPVTP---SAPPPSTPATPSSPPPSTTPSAPPPSTPSNSPPSP 179
Score = 32.0 bits (71), Expect = 0.74
Identities = 17/57 (29%), Positives = 22/57 (37%), Gaps = 1/57 (1%)
Frame = +3
Query: 572 TCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQ-PTEATQPPQPPTVAAQNP 627
T + PP S P+ P T A PP PP + P + P PP+ P
Sbjct: 129 TPSAPPPSTPSNSPPSPPTTPAISPPSGGGTTPSPPSRTPPSSDDSPSPPSSKTPPP 299
Score = 31.2 bits (69), Expect = 1.3
Identities = 18/54 (33%), Positives = 25/54 (45%)
Frame = +3
Query: 574 TNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
++PP + P+ P T A PP T +A PP P + PP T +A P
Sbjct: 3 SSPPPATPSSPPPST----PANPP-PVTPSAPPPSTPATPSSPPPSTTPSAPPP 149
Score = 30.8 bits (68), Expect = 1.6
Identities = 22/87 (25%), Positives = 29/87 (33%)
Frame = +3
Query: 534 RPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTCTNPPTSQPTEDVPETEPTEA 593
R KK RRRD +P R + PP P ++ +
Sbjct: 405 RKKKRRRRDEEYYGQQNYQQPPPAQR--------PKVEPYGGPPQQWQNNAHPPSDHVVS 560
Query: 594 AQPPQQPTEAAQPPQQPTEATQPPQPP 620
PP P PP+ P+ PP PP
Sbjct: 561 KPPPPAPI----PPRPPSHVAPPPPPP 629
Score = 28.9 bits (63), Expect = 6.2
Identities = 19/53 (35%), Positives = 22/53 (40%), Gaps = 6/53 (11%)
Frame = +3
Query: 581 PTEDVPETEPTEAAQPPQQPTEAAQPPQQ------PTEATQPPQPPTVAAQNP 627
P P+ EP PPQQ A PP P A PP+PP+ A P
Sbjct: 468 PPAQRPKVEPYGG--PPQQWQNNAHPPSDHVVSKPPPPAPIPPRPPSHVAPPP 620
>CB894755 similar to GP|9757890|db phosphoribosylanthranilate
transferase-like protein {Arabidopsis thaliana}, partial
(10%)
Length = 674
Score = 42.0 bits (97), Expect = 7e-04
Identities = 21/64 (32%), Positives = 30/64 (46%)
Frame = +3
Query: 564 GTIGHNIRTCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVA 623
G +G I +P+ VPE PT +++PP E + PP E +PP PP+ A
Sbjct: 315 GKVGLKIYYVDEEIPPRPSAPVPENPPTPSSEPPSVKVEESPPPPSEPEKVEPP-PPSEA 491
Query: 624 AQNP 627
P
Sbjct: 492 GPQP 503
>TC81858 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (9%)
Length = 748
Score = 41.2 bits (95), Expect = 0.001
Identities = 22/58 (37%), Positives = 28/58 (47%), Gaps = 1/58 (1%)
Frame = +1
Query: 574 TNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVA-AQNPVVS 630
T PP S T P ++ P PT A P + PTE+ P PTV+ A +PV S
Sbjct: 226 TTPPKSSATSPTSSPAPKVSSPPSPTPTSAEAPVESPTESPPAPVSPTVSPATSPVAS 399
>TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 - alfalfa,
partial (97%)
Length = 1049
Score = 40.0 bits (92), Expect = 0.003
Identities = 20/56 (35%), Positives = 25/56 (43%), Gaps = 1/56 (1%)
Frame = +3
Query: 576 PPTSQPTEDVPETEP-TEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNPVVS 630
PP Q P T P ++ PP P A+ PP P AT PP P A P ++
Sbjct: 429 PPVQQSPPPTPLTPPPVQSTPPPASPPPASPPPFSPPPATPPPATPPPATPPPALT 596
Score = 37.0 bits (84), Expect = 0.023
Identities = 19/57 (33%), Positives = 26/57 (45%), Gaps = 1/57 (1%)
Frame = +3
Query: 575 NPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPP-QPPTVAAQNPVVS 630
+PP + T ++ P A+ PP P + PP P AT PP PP P+ S
Sbjct: 444 SPPPTPLTPPPVQSTPPPASPPPASPPPFSPPPATPPPATPPPATPPPALTPTPLSS 614
Score = 35.4 bits (80), Expect = 0.067
Identities = 18/45 (40%), Positives = 19/45 (42%), Gaps = 1/45 (2%)
Frame = +3
Query: 574 TNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQ-QPTEATQPP 617
T PP S P P P A PP P A PP PT + PP
Sbjct: 486 TPPPASPPPASPPPFSPPPATPPPATPPPATPPPALTPTPLSSPP 620
Score = 32.3 bits (72), Expect = 0.56
Identities = 21/69 (30%), Positives = 31/69 (44%), Gaps = 12/69 (17%)
Frame = +3
Query: 574 TNPPTSQPTEDVPETEPTEAAQPPQQ----PTEAAQPPQQ----PTEATQPP----QPPT 621
T P + PT P +++ PP Q P +++ PP Q P +++ PP PPT
Sbjct: 279 TPPANTPPTTPQASPPPVQSSPPPVQSSPPPLQSSPPPAQSTPPPVQSSPPPVQQSPPPT 458
Query: 622 VAAQNPVVS 630
PV S
Sbjct: 459 PLTPPPVQS 485
Score = 32.0 bits (71), Expect = 0.74
Identities = 17/57 (29%), Positives = 22/57 (37%), Gaps = 2/57 (3%)
Frame = +3
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPP--QQPTEATQPPQPPTVAAQNPVVS 630
PP S P P P A PP P A P P T P P + ++ P ++
Sbjct: 507 PPASPPPFSPPPATPPPATPPPATPPPALTPTPLSSPPATTPAPAPAKLKSKAPALA 677
Score = 30.0 bits (66), Expect = 2.8
Identities = 17/59 (28%), Positives = 25/59 (41%), Gaps = 4/59 (6%)
Frame = +3
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQ----PTEAAQPPQQPTEATQPPQPPTVAAQNPVVS 630
P S P P +++ PP Q P +++ PP Q + P PP V + P S
Sbjct: 327 PVQSSPPPVQSSPPPLQSSPPPAQSTPPPVQSSPPPVQQSPPPTPLTPPPVQSTPPPAS 503
Score = 30.0 bits (66), Expect = 2.8
Identities = 20/77 (25%), Positives = 31/77 (39%), Gaps = 7/77 (9%)
Frame = +3
Query: 560 CGRCGTIGHNIRTCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQ----PTEATQ 615
C ++G T+P +S P P Q P +++ PP Q P +++
Sbjct: 207 CVVFSSVGAQQAPSTSPNSSPAPPTPPANTPPTTPQASPPPVQSSPPPVQSSPPPLQSSP 386
Query: 616 PP---QPPTVAAQNPVV 629
PP PP V + P V
Sbjct: 387 PPAQSTPPPVQSSPPPV 437
Score = 29.6 bits (65), Expect = 3.7
Identities = 18/65 (27%), Positives = 27/65 (40%), Gaps = 11/65 (16%)
Frame = +3
Query: 574 TNPPTSQPTEDVPETEPTEAAQPPQQ-----------PTEAAQPPQQPTEATQPPQPPTV 622
++PP +Q T P +++ PP Q P ++ PP P A+ PP P
Sbjct: 378 SSPPPAQSTPP-----PVQSSPPPVQQSPPPTPLTPPPVQSTPPPASPPPASPPPFSPPP 542
Query: 623 AAQNP 627
A P
Sbjct: 543 ATPPP 557
Score = 29.3 bits (64), Expect = 4.8
Identities = 16/54 (29%), Positives = 20/54 (36%)
Frame = +3
Query: 574 TNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
+ PP + P P P + PP P A PP P A P + A P
Sbjct: 483 STPPPASP----PPASPPPFSPPPATPPPATPPPATPPPALTPTPLSSPPATTP 632
>BF638163
Length = 687
Score = 38.5 bits (88), Expect = 0.008
Identities = 21/70 (30%), Positives = 32/70 (45%)
Frame = +2
Query: 559 RCGRCGTIGHNIRTCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQ 618
+CGRCG +GH+ R+C ++ ++ +TEP E Q E AT+ Q
Sbjct: 404 QCGRCGLMGHSSRSCAK*GVARRPKNWIDTEPEEQVQSNVASVETV----VENVATEVQQ 571
Query: 619 PPTVAAQNPV 628
V NP+
Sbjct: 572 EDVVRVSNPI 601
>TC82364 similar to GP|20268740|gb|AAM14073.1 unknown protein {Arabidopsis
thaliana}, partial (39%)
Length = 1174
Score = 38.5 bits (88), Expect = 0.008
Identities = 19/55 (34%), Positives = 26/55 (46%)
Frame = +3
Query: 574 TNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNPV 628
+NP QP P + P + PP +P QP+ TQPP PP A+ P+
Sbjct: 123 SNPLFQQPIPIQPVSYPVKPQDPPPP-----EPESQPSSDTQPPPPPPQASNTPI 272
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 38.1 bits (87), Expect = 0.010
Identities = 41/145 (28%), Positives = 48/145 (32%), Gaps = 24/145 (16%)
Frame = +3
Query: 507 TPINGKRMWPYTPDAPILPPLYKRKPGR-----PKKLRRRDPHEDDTD--------EGS- 552
TP + R P+TP P PP + P R P R P DD EG
Sbjct: 228 TPTHPPR--PHTPPTP--PPTTQTTPPREAPTDPGTTRGTPPTADDAQTSPAHHPGEGGG 395
Query: 553 ---------RPNSRH-RCGRCGTIGHNIRTCTNPPTSQPTEDVPETEPTEAAQPPQQPTE 602
RP RH R G RT T P Q D P T+P + PP E
Sbjct: 396 KGGAPPPPPRPPPRHGRGGHQNQTREAKRTTTEP---QGKRDRPRTDPRKKKHPPPHQKE 566
Query: 603 AAQPPQQPTEATQPPQPPTVAAQNP 627
++P PP P P
Sbjct: 567 GGNTQKEPRGEDPPPHTPAGGGGGP 641
Score = 36.2 bits (82), Expect = 0.039
Identities = 31/112 (27%), Positives = 36/112 (31%), Gaps = 7/112 (6%)
Frame = +1
Query: 516 PYTPDAPILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTCTN 575
P TP PP KR P R E+ T + R + + N
Sbjct: 4 PTTPQ----PPAPKRPPKEKDTQARNPQPEEATKKQKRAPEKKK---------------N 126
Query: 576 PPTSQPTEDVPETEPTEAAQPP--QQPTEAAQP-----PQQPTEATQPPQPP 620
PP QP P +PP Q P P P PT PPQPP
Sbjct: 127 PPRGQPPPPPPPPPQAPPTKPPTRQTPKNNTPPPPPHTPPDPTPPQPPPQPP 282
Score = 31.2 bits (69), Expect = 1.3
Identities = 19/61 (31%), Positives = 23/61 (37%), Gaps = 4/61 (6%)
Frame = +3
Query: 571 RTCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPP----QPPTVAAQN 626
R T PP P +T T + PT PP+ T T PP PP A +
Sbjct: 135 RPATPPPAPAPPGPPNQTTNTTNPKKQHTPTTPTHPPRPHTPPTPPPTTQTTPPREAPTD 314
Query: 627 P 627
P
Sbjct: 315 P 317
Score = 30.8 bits (68), Expect = 1.6
Identities = 28/110 (25%), Positives = 37/110 (33%), Gaps = 11/110 (10%)
Frame = +2
Query: 516 PYTPDAPILPPLYKRKPGRPKKLRRRDPHEDDTD---------EGSRPNSRHRCGRCGTI 566
P P P PP K++ +P+ R+ P ++ + E S P R R R
Sbjct: 11 PPNPQPPSDPP--KKRTPKPETPNRKKPQKNKKEPPKKRKTPPEASHPPPRPRPPR---- 172
Query: 567 GHNIRTCTNPPTSQPTEDVPE--TEPTEAAQPPQQPTEAAQPPQQPTEAT 614
PP D P+ T P PP P PP P T
Sbjct: 173 ---------PPQPNHQHDKPQKTTHPHHPHTPPPTPHPPNPPPNHPNHPT 295
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 37.0 bits (84), Expect = 0.023
Identities = 26/78 (33%), Positives = 32/78 (40%), Gaps = 3/78 (3%)
Frame = -3
Query: 553 RPNS--RHRCGRCGTIGHNIRTCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQP-PQQ 609
RP+S + CGR GH PP +P P P +PP P + P P
Sbjct: 255 RPHSP*QAACGR----GH-------PPRHRPPPPGPPPPPPLGPRPPPHPPRSGPPRPHP 109
Query: 610 PTEATQPPQPPTVAAQNP 627
PT + P PP AA P
Sbjct: 108 PTRTSLSPPPPPPAASPP 55
Score = 35.8 bits (81), Expect = 0.051
Identities = 34/125 (27%), Positives = 42/125 (33%), Gaps = 3/125 (2%)
Frame = -3
Query: 505 GITPINGKRMWPYTPDAPILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCG 564
G TP G R P +P P + G P + R P + G P RHR G
Sbjct: 351 GGTP--GSRAPSPAPPSPTHPAPRRPWTGPPPRPRPHSP*QAACGRGHPP--RHRPPPPG 184
Query: 565 TIGHNIRTCTNPPTSQPTEDVPETEPTEAAQPPQQP---TEAAQPPQQPTEATQPPQPPT 621
PP P P P + P P T + PP P + PP PP
Sbjct: 183 -----------PPPPPPLGPRPPPHPPRSGPPRPHPPTRTSLSPPPPPPAASPPPPAPPP 37
Query: 622 VAAQN 626
+ N
Sbjct: 36 LLVYN 22
Score = 35.0 bits (79), Expect = 0.087
Identities = 32/115 (27%), Positives = 43/115 (36%), Gaps = 3/115 (2%)
Frame = -3
Query: 516 PYTPDAPILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTCTN 575
P P P PP +P + P G+RP+ R I T+
Sbjct: 690 PPPPPPPSSPPGMGTRPSCQENWWAAPP-------GARPSRPSRY---------IPPPTS 559
Query: 576 PPTSQPTEDVPETEPTEA---AQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
PP + P VP P+ A PP P +PP +P A Q P+P A+ P
Sbjct: 558 PPPASPPAPVPPQSPSTIRGRAPPPAPPPR--RPPARPPPAPQRPRPRRPPARPP 400
Score = 34.7 bits (78), Expect = 0.11
Identities = 31/127 (24%), Positives = 42/127 (32%), Gaps = 18/127 (14%)
Frame = -1
Query: 519 PDAPILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTCTNPPT 578
P AP+ PP G P+ R+ G+ P + HR + T PP
Sbjct: 461 PRAPLRPPSAPDPAGHPRAPRQGVAVVPGPRSGNSPRAAHRGPEPRPLPRPPPP-TRPPA 285
Query: 579 SQPTEDVPETEPTEAAQPP------------------QQPTEAAQPPQQPTEATQPPQPP 620
+ P PT ++PP + P+ A PP P A P PP
Sbjct: 284 APGPGPPPAPGPTPLSKPPAAEGIPPATAPPPRARRLRPPSAPAPPPTPPAAAPPAPTPP 105
Query: 621 TVAAQNP 627
A P
Sbjct: 104 HAPA*AP 84
Score = 30.8 bits (68), Expect = 1.6
Identities = 18/54 (33%), Positives = 22/54 (40%)
Frame = -2
Query: 577 PTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNPVVS 630
P + P P P +A PP+ P PPQ+P PP PP P S
Sbjct: 223 PRASPPPPPPPPGPAASA-PPRPPPPPPPPPQRPP----PPPPPHTHQPEPPTS 77
Score = 30.4 bits (67), Expect = 2.1
Identities = 19/58 (32%), Positives = 19/58 (32%), Gaps = 6/58 (10%)
Frame = -2
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPP----QQPTEATQPP--QPPTVAAQNP 627
PP P P PP P PP QP T PP QPP A P
Sbjct: 208 PPPPPPPGPAASAPPRPPPPPPPPPQRPPPPPPPHTHQPEPPTSPPRRQPPAPRAPPP 35
Score = 30.4 bits (67), Expect = 2.1
Identities = 30/113 (26%), Positives = 37/113 (32%)
Frame = -3
Query: 508 PINGKRMWPYTPDAPILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIG 567
P R P P P +R P RP RRR G+R R + + GT G
Sbjct: 486 PAPPPRRPPARPPPAPQRPRPRRPPARPPSGRRR---------GARAKVR-KLPKGGTPG 337
Query: 568 HNIRTCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPP 620
+ P + P P T P +P A P PP PP
Sbjct: 336 SRAPSPAPPSPTHPAPRRPWTGPPPRPRPHSP*QAACGRGHPPRHRPPPPGPP 178
>BG457880 weakly similar to GP|1063689|gb|A ORF 66; this sequence overlaps at
the HindIII site with the sequence reported for the
OpMNPV lef-3, partial (5%)
Length = 669
Score = 37.0 bits (84), Expect = 0.023
Identities = 36/125 (28%), Positives = 49/125 (38%), Gaps = 14/125 (11%)
Frame = +3
Query: 516 PYTPDAPILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPN----SRHRCGRCGTIGHNIR 571
P P A Y P +P++ P DT++ PN S++ G
Sbjct: 120 PQVPTASDPNSSYYLTPHQPQQTPL-GPSAPDTNQSPYPNLQQPSQYPGGSVSAPSQPTP 296
Query: 572 TCTNPPTSQPTEDVPE-----TEPTEAAQP-PQQPTEAAQPPQQPT----EATQPPQPPT 621
T +QP + V +P + +QP P QPT+ A PPQQ Q Q P
Sbjct: 297 GQTPAQVAQPVQPVQSPHMQHAQPPQPSQPQPPQPTQQAPPPQQQAYWQPSMPQSHQQPP 476
Query: 622 VAAQN 626
VA QN
Sbjct: 477 VAPQN 491
Score = 30.0 bits (66), Expect = 2.8
Identities = 15/41 (36%), Positives = 21/41 (50%)
Frame = +3
Query: 586 PETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQN 626
P+ + QPPQQP + P T+ +Q PQ PT + N
Sbjct: 39 PQDPYQQQQQPPQQPND----PNLQTQNSQQPQVPTASDPN 149
>BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania major}, partial
(1%)
Length = 766
Score = 37.0 bits (84), Expect = 0.023
Identities = 23/72 (31%), Positives = 28/72 (37%), Gaps = 13/72 (18%)
Frame = +1
Query: 569 NIRTCTNPPTSQPTEDVPETEPTEAAQ-PPQQPTEAAQP------------PQQPTEATQ 615
N+ T PP S P PT A+ PP PT + P P PT
Sbjct: 25 NLHTTWQPPASSSPSARPSPPPTTASSTPPGAPTPSPTPTRRATLNGNTPAPASPTAQAT 204
Query: 616 PPQPPTVAAQNP 627
PP P+ AA +P
Sbjct: 205 PPTGPSTAAPSP 240
Score = 28.9 bits (63), Expect = 6.2
Identities = 21/80 (26%), Positives = 28/80 (34%), Gaps = 3/80 (3%)
Frame = +1
Query: 548 TDEGSRPNSRHRCGRCGTIGHNIRTCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPP 607
T R +R R R CT PTS + PP P AA+P
Sbjct: 382 TSRPPRARARRAASRSSPWATPARACTTAPTSASRQ-----------TPPPSPRTAAKPK 528
Query: 608 QQ---PTEATQPPQPPTVAA 624
P+ + P +P +AA
Sbjct: 529 ASRCTPSRSRAPTEPSKMAA 588
>AJ498355
Length = 661
Score = 36.6 bits (83), Expect = 0.030
Identities = 36/143 (25%), Positives = 60/143 (41%), Gaps = 8/143 (5%)
Frame = +1
Query: 408 NHPFMPKPLKRLAWETKSSNNWVPQACGHAKYEVKSVL------DGDQFIVDLRQRSCTC 461
N+ P ++L ++ K+ VP K EVKS + D + +DL QR
Sbjct: 115 NNDSKPPQRRKLKFQPKAPPRRVP------KVEVKSEVIEEDNKDVSKVTMDLLQRHNEN 276
Query: 462 NWWDLVGIPCRHAVAAISFNGGHAESYIDSY--YGRDAYQATYAHGITPINGKRMWPYTP 519
+ + + I+F G +Y+ SY G + + +G+T K W
Sbjct: 277 AMNRRMKHEKKVLPSQIAFGAGGQSTYLKSYGTKGGSKSENSAFYGVTEKEYKEPWELDT 456
Query: 520 DAPILPPLYKRKPGRPKKLRRRD 542
+ PI+ PL K G P+KL ++
Sbjct: 457 EYPIVHPLRKPYSGNPEKLDEKE 525
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.137 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,305,176
Number of Sequences: 36976
Number of extensions: 400777
Number of successful extensions: 5885
Number of sequences better than 10.0: 177
Number of HSP's better than 10.0 without gapping: 2955
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4543
length of query: 630
length of database: 9,014,727
effective HSP length: 102
effective length of query: 528
effective length of database: 5,243,175
effective search space: 2768396400
effective search space used: 2768396400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0137.1


