

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0135.9
(645 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
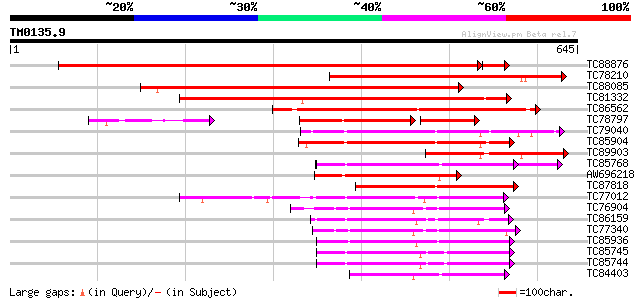
Score E
Sequences producing significant alignments: (bits) Value
TC88876 similar to PIR|G96537|G96537 hypothetical protein F2J10.... 813 0.0
TC78210 similar to GP|6056413|gb|AAF02877.1| Unknown protein {Ar... 346 1e-95
TC88085 similar to GP|20259341|gb|AAM13995.1 unknown protein {Ar... 340 1e-93
TC81332 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22... 328 5e-90
TC86562 similar to GP|21553404|gb|AAM62497.1 26S proteasome regu... 274 7e-74
TC78797 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22... 173 4e-64
TC79040 homologue to PIR|F84674|F84674 probable AAA-type ATPase ... 196 2e-50
TC85904 similar to PIR|F84674|F84674 probable AAA-type ATPase [i... 191 6e-49
TC89903 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22... 180 1e-45
TC85768 homologue to SP|P54774|CC48_SOYBN Cell division cycle pr... 172 4e-43
AW696218 homologue to PIR|B96835|B96 CAD ATPase (AAA1) 35570-33... 166 3e-41
TC87818 similar to GP|20197082|gb|AAC26698.2 putative katanin {A... 151 9e-37
TC77012 homologue to SP|P54776|PRSA_LYCES 26S protease regulator... 150 2e-36
TC76904 homologue to GP|4325041|gb|AAD17230.1| FtsH-like protein... 147 2e-35
TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AA... 146 2e-35
TC77340 SP|Q9BAE0|FTSH_MEDSA Cell division protein ftsH homolog ... 144 1e-34
TC85936 homologue to GP|13537115|dbj|BAB40755. AtSUG1 {Arabidops... 144 1e-34
TC85745 homologue to GP|17297987|dbj|BAB78491. 26S proteasome re... 140 1e-33
TC85744 homologue to PIR|T08959|T08959 proteinase homolog F19B15... 139 3e-33
TC84403 homologue to GP|15021761|gb|AAK77908.1 AAA-metalloprotea... 134 1e-31
>TC88876 similar to PIR|G96537|G96537 hypothetical protein F2J10.1
[imported] - Arabidopsis thaliana, partial (72%)
Length = 1572
Score = 813 bits (2101), Expect(2) = 0.0
Identities = 405/484 (83%), Positives = 446/484 (91%), Gaps = 2/484 (0%)
Frame = +2
Query: 56 RENEVELESVNNDGRKPPVYWINVKNIEHDLDAQSQDCYLAMEALREVLNSKQPLIVYFP 115
R ++ E++NN KP VYWINVK+IE+DLDAQS DCY+A+EAL EVLNSK+PLIVYFP
Sbjct: 29 RPMKMRWENLNNSRAKPSVYWINVKDIENDLDAQSHDCYIAVEALCEVLNSKRPLIVYFP 208
Query: 116 DGSQWLHKSVPKSIRSEFFHKVEEMFDQLSGPIVLICGQNKAQSGTKEKGQFTMILPNFA 175
D SQWLHKSVPKS R+EFFHKVEEMFD+L GP+VLICGQNK SG+KEK +FTMILPNF
Sbjct: 209 DSSQWLHKSVPKSNRNEFFHKVEEMFDRLYGPVVLICGQNKVHSGSKEKEKFTMILPNFG 388
Query: 176 RLAKLPLSLKRLTD--KGQKTSDDDEINKLFSNVVSIQPPKDENLLATFKKQLEEDRKIV 233
R+AKLPLSLK LTD KG KTS++D+INKLFSNV+S+ PPK+ENL FKKQLEEDRKIV
Sbjct: 389 RVAKLPLSLKHLTDGFKGGKTSEEDDINKLFSNVLSVHPPKEENLQTVFKKQLEEDRKIV 568
Query: 234 ISRSNLNELRKVLEEHQLSCVDLLHLDSDDVILTKQKAEKVVGWAKNHYLSSCLLPCVKG 293
ISRSNLNELRKVLEEHQLSC DLLH+++D +++TKQKAEK+VGWAKNHYLSSCLLP +KG
Sbjct: 569 ISRSNLNELRKVLEEHQLSCTDLLHVNTDGIVITKQKAEKLVGWAKNHYLSSCLLPSIKG 748
Query: 294 DKLCLPRESLEIAISRLKGVETMSRQPSQSLKNLAKDEFESNFISAVVPPGEIGVKFDDI 353
++LC+PRESLEIAISR+KG+ETMSR+ SQ+LKNLAKDEFESNF+SAVV PGEIGVKFDDI
Sbjct: 749 ERLCIPRESLEIAISRMKGMETMSRKSSQNLKNLAKDEFESNFVSAVVAPGEIGVKFDDI 928
Query: 354 GALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGAN 413
GALEDVKKAL ELVILPMRRPELF+HGNLLRP KGILLFGPPGTGKTL+AKALATEAGAN
Sbjct: 929 GALEDVKKALQELVILPMRRPELFSHGNLLRPCKGILLFGPPGTGKTLLAKALATEAGAN 1108
Query: 414 FISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRM 473
FISV+GSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGA EHEATRRM
Sbjct: 1109FISVTGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAHEHEATRRM 1288
Query: 474 RNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRII 533
RNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPD +NR KILRI
Sbjct: 1289RNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDVENRMKILRIF 1468
Query: 534 LKHE 537
L E
Sbjct: 1469LAKE 1480
Score = 58.9 bits (141), Expect(2) = 0.0
Identities = 25/31 (80%), Positives = 30/31 (96%)
Frame = +1
Query: 538 NLDPDFQFDKLANLTDGYSGSDLKNLCITAA 568
NL+PDFQ+DKLA+LT+GYSGSDLKNLC+ AA
Sbjct: 1480 NLNPDFQYDKLASLTEGYSGSDLKNLCVAAA 1572
>TC78210 similar to GP|6056413|gb|AAF02877.1| Unknown protein {Arabidopsis
thaliana}, partial (23%)
Length = 1254
Score = 346 bits (888), Expect = 1e-95
Identities = 169/280 (60%), Positives = 221/280 (78%), Gaps = 11/280 (3%)
Frame = +3
Query: 365 ELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTS 424
ELV+LP++RPELF G L +P KGILLFGPPGTGKT++AKA+ATEAGANFI++S S++TS
Sbjct: 6 ELVMLPLKRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITS 185
Query: 425 KWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGL 484
KWFG+ EK KA+FS ASK+AP +IFVDEVDS+LG R EHEA R+M+NEFM WDGL
Sbjct: 186 KWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGL 365
Query: 485 RSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQ 544
R+KE +RIL+L ATNRPFDLD+AVIRRLPRR+ VDLPDA NR KILR+IL E+L D
Sbjct: 366 RTKEKERILVLAATNRPFDLDEAVIRRLPRRLMVDLPDAPNRGKILRVILAKEDLAADVD 545
Query: 545 FDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEK------AGDNN-----ITNVALRP 593
+ +AN+TDGYSGSDLKNLC+TAA+ P++E+LE+EK +N ++ +RP
Sbjct: 546 LEAIANMTDGYSGSDLKNLCVTAAHCPIREILEKEKKDKSLALAENKPEPELCSSADIRP 725
Query: 594 LNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTR 633
L ++DF + +V SV+ ++ ++NEL++WN++YGEGG+R
Sbjct: 726 LKMEDFRYAHEQVCASVSSESTNMNELQQWNDLYGEGGSR 845
>TC88085 similar to GP|20259341|gb|AAM13995.1 unknown protein {Arabidopsis
thaliana}, partial (29%)
Length = 1261
Score = 340 bits (872), Expect = 1e-93
Identities = 181/377 (48%), Positives = 255/377 (67%), Gaps = 9/377 (2%)
Frame = +1
Query: 149 VLICGQNKAQSGTKEKGQ-----FTMILPNFARLAKL--PLSLKRLTDKGQKTSDD-DEI 200
V++ G + KEK Q FT N L L P + RL D+ ++T ++
Sbjct: 130 VVVIGSHIHPDNRKEKTQPGSLLFTKFGGNQTALLDLAFPDNFTRLHDRSKETPKVMKQL 309
Query: 201 NKLFSNVVSIQPPKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLD 260
N+ F N V+IQ P+DE LL+ +K+ LE D + + ++SN+ +R VL + L C +L L
Sbjct: 310 NRFFPNKVTIQLPQDEALLSDWKQHLERDVETMKAQSNVVSIRLVLNKFGLDCPELETLS 489
Query: 261 SDDVILTKQKAEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAISRLKGVETMSRQP 320
D LT + EK++GWA +++ + K + ES++ + L+G++ ++
Sbjct: 490 IKDQTLTTENVEKIIGWAISYHFMHSSEASTEESKPVISAESIQYGFNILQGIQNENKSV 669
Query: 321 SQSLKNLA-KDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTH 379
+SLK++ ++EFE + V+PP +IGV F+DIGALE+VK L ELV+LP++RPELF
Sbjct: 670 KKSLKDVVTENEFEKKLLGDVIPPTDIGVSFNDIGALENVKDTLKELVMLPLQRPELFCK 849
Query: 380 GNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFS 439
G L +P KGILLFGPPGTGKT++AKA+ATEAGANFI++S S++TSKWFG+ EK KA+FS
Sbjct: 850 GQLTKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGEKYVKAVFS 1029
Query: 440 FASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATN 499
ASK+AP +IFVDEVDS+LG R EHEA R+M+NEFM WDGLR+K+ +R+L+L ATN
Sbjct: 1030LASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDRERVLVLAATN 1209
Query: 500 RPFDLDDAVIRRLPRRI 516
RPFDLD+AVIRRLPRR+
Sbjct: 1210RPFDLDEAVIRRLPRRL 1260
>TC81332 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22
{Arabidopsis thaliana}, partial (39%)
Length = 1514
Score = 328 bits (840), Expect = 5e-90
Identities = 177/405 (43%), Positives = 251/405 (61%), Gaps = 27/405 (6%)
Frame = +1
Query: 194 TSDDDEINKLFSNVVSIQPPKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSC 253
T ++++ LF + I PP+DE L +K QL++ K + + +VL + L C
Sbjct: 286 TKVNEKLTMLFPYNIEITPPQDETHLKIWKSQLKKAMKKTHLKDYTTHIAEVLAANDLYC 465
Query: 254 VDLLHLDSDDVILTKQKAEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAISRLKGV 313
DL +D +D+ + + E+VV A H+L P + L + +SL +S +
Sbjct: 466 DDLDTVDHNDMTILSNQTEEVVASAIFHHLKDAKNPKYRNGILIISAKSLRHVLSLFQEG 645
Query: 314 ETMSRQPSQSLKNLAKDE---------------------------FESNFISAVVPPGEI 346
E+ + ++ K +D+ FE ++P EI
Sbjct: 646 ESSEKDNKKTKKESKRDDSRKEKPKESKKDGDIKASAKSDSPDNAFEECIRQELIPANEI 825
Query: 347 GVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKAL 406
V F DIGAL+DVK++L E V+LP+RRP+LF +L+P KG+LLFGPPGTGKT++AKA+
Sbjct: 826 KVTFSDIGALDDVKESLQEAVMLPLRRPDLFKGDGVLKPCKGVLLFGPPGTGKTMLAKAI 1005
Query: 407 ATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFE 466
A EAGA+FI+VS ST+TS W G +EK +ALFS A+K+AP IIF+DEVDS+LG R E
Sbjct: 1006ANEAGASFINVSPSTITSMWQGQSEKNVRALFSLAAKVAPTIIFIDEVDSMLGQRSSTRE 1185
Query: 467 HEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNR 526
H + RR++NEFM+ WDGL SK +++I +L ATN PFDLD+A+IRR RRI V LP ADNR
Sbjct: 1186HSSMRRVKNEFMSRWDGLLSKPDEKITVLAATNMPFDLDEAIIRRFQRRIMVGLPSADNR 1365
Query: 527 KKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRP 571
+ IL+ IL E + + F++L+ +T+GYSGSDLKNLC+TAAYRP
Sbjct: 1366ETILKTILAKEKSE-NMNFEELSTMTEGYSGSDLKNLCMTAAYRP 1497
>TC86562 similar to GP|21553404|gb|AAM62497.1 26S proteasome regulatory
particle chain RPT6-like protein {Arabidopsis thaliana},
partial (77%)
Length = 1564
Score = 274 bits (701), Expect = 7e-74
Identities = 144/305 (47%), Positives = 201/305 (65%)
Frame = +1
Query: 300 RESLEIAISRLKGVETMSRQPSQSLKNLAKDEFESNFISAVVPPGEIGVKFDDIGALEDV 359
RES + A+ + K + +P + + +E V+ P I V+FD IG LE +
Sbjct: 268 RESSKKALEQKKEIAKRLGRPL-----IQTNSYEDVIACDVINPDHIDVEFDSIGGLETI 432
Query: 360 KKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSG 419
K+ L ELVILP++RP+LF+HG LL P KG+LL+GPPGTGKT++AKA+A E+GA FI+V
Sbjct: 433 KQTLFELVILPLQRPDLFSHGKLLGPQKGVLLYGPPGTGKTMLAKAIAKESGAVFINVRI 612
Query: 420 STLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMA 479
S L SKWFGDA+KL A+FS A KL P IIF+DEVDS LG R + +HEA M+ EFMA
Sbjct: 613 SNLMSKWFGDAQKLVAAVFSLAHKLQPSIIFIDEVDSFLGQRRSS-DHEAVLNMKTEFMA 789
Query: 480 AWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENL 539
WDG + ++ R+++L ATNRP +LD+A++RRLP+ + PD R IL++ILK E +
Sbjct: 790 LWDGFATDQSARVMVLAATNRPSELDEAILRRLPQAFEIGYPDRKERADILKVILKGEKV 969
Query: 540 DPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITNVALRPLNLDDF 599
+ + F +A L GY+GSDL +LC AAY P++E+L EK G+ + RPL+ D
Sbjct: 970 EDNIDFSYIAGLCKGYTGSDLFDLCKKAAYFPIREILHNEKNGEQSCEP---RPLSQADL 1140
Query: 600 VQSKS 604
++ S
Sbjct: 1141EKALS 1155
>TC78797 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22 {Arabidopsis
thaliana}, partial (74%)
Length = 2270
Score = 173 bits (439), Expect(2) = 4e-64
Identities = 86/132 (65%), Positives = 108/132 (81%)
Frame = +1
Query: 330 DEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGI 389
+EFE V+P EI V F DIGALE+ K++L ELV+LP+RRP+LFT G LL+P +GI
Sbjct: 1651 NEFEKRIRPEVIPANEIDVTFSDIGALEETKESLQELVMLPLRRPDLFT-GGLLKPCRGI 1827
Query: 390 LLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVII 449
LLFGPPGTGKT++AKA+A EAGA+FI+VS ST+TSKWFG+ EK +ALF+ ASK++P II
Sbjct: 1828 LLFGPPGTGKTMLAKAIAKEAGASFINVSMSTITSKWFGEDEKNVRALFTLASKVSPTII 2007
Query: 450 FVDEVDSLLGAR 461
FVDEVDS+LG R
Sbjct: 2008 FVDEVDSMLGQR 2043
Score = 90.5 bits (223), Expect(2) = 4e-64
Identities = 40/67 (59%), Positives = 55/67 (81%)
Frame = +3
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRK 527
+A R+++NEFM+ WDGL +K+ +RIL+L ATN+PFDLD+A+IRR RRI V LP +NR+
Sbjct: 2064 KAMRKIKNEFMSHWDGLTTKQGERILVLAATNKPFDLDEAIIRRFERRIMVGLPSVENRE 2243
Query: 528 KILRIIL 534
KILR +L
Sbjct: 2244 KILRTLL 2264
Score = 45.8 bits (107), Expect = 5e-05
Identities = 41/147 (27%), Positives = 68/147 (45%), Gaps = 3/147 (2%)
Frame = +2
Query: 90 SQDCYLAMEALREVLNSKQ---PLIVYFPDGSQWLHKSVPKSIRSEFFHKVEEMFDQLSG 146
S D L +++L +VL S P+++Y D + L +S + ++ ++M +LSG
Sbjct: 851 SFDEKLLIQSLYKVLLSVSKTYPIVLYLRDVDRLLSRS------QKIYNMFQKMLKRLSG 1012
Query: 147 PIVLICGQNKAQSGTKEKGQFTMILPNFARLAKLPLSLKRLTDKGQKTSDDDEINKLFSN 206
PI LI G SG + F L D+++ LF
Sbjct: 1013PI-LILGSRVLDSGNE-----------FEEL-------------------DEKLTVLFPY 1099
Query: 207 VVSIQPPKDENLLATFKKQLEEDRKIV 233
+ I+PP+DE+ L ++K QLEED K++
Sbjct: 1100DIKIRPPEDESHLVSWKSQLEEDMKMI 1180
>TC79040 homologue to PIR|F84674|F84674 probable AAA-type ATPase [imported]
- Arabidopsis thaliana, partial (74%)
Length = 1193
Score = 196 bits (498), Expect = 2e-50
Identities = 118/331 (35%), Positives = 188/331 (56%), Gaps = 30/331 (9%)
Frame = +3
Query: 331 EFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGIL 390
+ + ISA++ + VK++D+ LE K+AL E VILP++ P+ FT RP + L
Sbjct: 3 KLRAGLISAIIRE-KPNVKWNDVAGLESAKQALQEAVILPVKFPQFFTGKR--RPWRAFL 173
Query: 391 LFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIF 450
L+GPPGTGK+ +AKA+ATEA + F S+S S L SKW G++EKL LF A + AP IIF
Sbjct: 174 LYGPPGTGKSYLAKAVATEADSTFFSISSSDLVSKWMGESEKLVSNLFQMARESAPSIIF 353
Query: 451 VDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR 510
VDE+DSL G RG E EA+RR++ E + G+ +Q++L+L ATN P+ LD A+ R
Sbjct: 354 VDEIDSLCGQRGEGNESEASRRIKTELLVQMQGV-GNNDQKVLVLAATNTPYALDQAIRR 530
Query: 511 RLPRRIYVDLPDADNRKKILRIIL---KHENLDPDFQFDKLANLTDGYSGSDLKNLCITA 567
R +RIY+ LPD R+ + ++ L H + D+++ LA+ T+G+SGSD+
Sbjct: 531 RFDKRIYIPLPDLKARQHMFKVHLGDTPHNLTEKDYEY--LASRTEGFSGSDISVCVKDV 704
Query: 568 AYRPVQELLE------------------EEKAGDNNITNVALR---------PLNLDDFV 600
+ PV++ + ++ A +T++A + P+ DF
Sbjct: 705 LFEPVRKTQDAMFFFKSPEGMWIPCGPKQQGAVQTTMTDLATKGLASKILPPPITRTDFE 884
Query: 601 QSKSKVGPSVAYDAVSVNELRKWNEMYGEGG 631
+ ++ P+V+ + V+E ++ + +GE G
Sbjct: 885 KVLARQRPTVSKSDLEVHE--RFTKEFGEEG 971
>TC85904 similar to PIR|F84674|F84674 probable AAA-type ATPase [imported] -
Arabidopsis thaliana, complete
Length = 1793
Score = 191 bits (486), Expect = 6e-49
Identities = 105/252 (41%), Positives = 156/252 (61%), Gaps = 6/252 (2%)
Frame = +2
Query: 329 KDEFESNF---ISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRP 385
+D ++S +++ + + VK++D+ LE K++L E VILP++ P+ FT RP
Sbjct: 503 RDAYQSKLRAGLNSAIVREKPNVKWNDVAGLESAKQSLQEAVILPVKFPQFFTGKR--RP 676
Query: 386 VKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLA 445
+ LL+GPPGTGK+ +AKA+ATEA + F SVS S L SKW G++EKL LF A + A
Sbjct: 677 WRAFLLYGPPGTGKSYLAKAVATEADSTFFSVSSSDLVSKWMGESEKLVSNLFEMARESA 856
Query: 446 PVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLD 505
P IIFVDE+DSL G RG E EA+RR++ E + G+ +Q++L+L ATN P+ LD
Sbjct: 857 PSIIFVDEIDSLCGTRGEGNESEASRRIKTELLVQMQGV-GHNDQKVLVLAATNTPYALD 1033
Query: 506 DAVIRRLPRRIYVDLPDADNRKKILRIIL---KHENLDPDFQFDKLANLTDGYSGSDLKN 562
A+ RR +RIY+ LPD R+ + ++ L H + D F+ LA T+G+SGSD+
Sbjct: 1034QAIRRRFDKRIYIPLPDLKARQHMFKVHLGDTPHNLTESD--FEHLARKTEGFSGSDIAV 1207
Query: 563 LCITAAYRPVQE 574
+ PV++
Sbjct: 1208CVKDVLFEPVRK 1243
>TC89903 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22
{Arabidopsis thaliana}, partial (20%)
Length = 875
Score = 180 bits (457), Expect = 1e-45
Identities = 93/186 (50%), Positives = 128/186 (68%), Gaps = 24/186 (12%)
Frame = +1
Query: 474 RNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRII 533
+NEFM WDGL S N++IL+LGATNRPFDLD+A+IRR RRI V LP A+NR+ IL+ +
Sbjct: 1 KNEFMTHWDGLLSGPNEQILVLGATNRPFDLDEAIIRRFERRIMVGLPSAENREMILKTL 180
Query: 534 L---KHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEK---------- 580
+ KHENLD F +LA +T+G+SGSDLKNLCITAAYRPV+EL+++E+
Sbjct: 181 MSKEKHENLD----FKELATMTEGFSGSDLKNLCITAAYRPVRELIQQERKKEMEKKKKE 348
Query: 581 -----------AGDNNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGE 629
+ DN+ + LRPL+++D Q+K++V S A + + EL++WNE+YGE
Sbjct: 349 AESVNSEDASNSNDNDEPEIILRPLSMEDMKQAKNQVAASFASEGSVMAELKQWNELYGE 528
Query: 630 GGTRTK 635
GG+R K
Sbjct: 529 GGSRKK 546
>TC85768 homologue to SP|P54774|CC48_SOYBN Cell division cycle protein 48
homolog (Valosin containing protein homolog) (VCP).
[Soybean], partial (95%)
Length = 2785
Score = 172 bits (436), Expect = 4e-43
Identities = 93/234 (39%), Positives = 140/234 (59%), Gaps = 4/234 (1%)
Frame = +2
Query: 350 FDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATE 409
+DDIG LE+VK+ L E V P+ PE F + P KG+L +GPPG GKTL+AKA+A E
Sbjct: 1532 WDDIGGLENVKRELQETVQYPVEHPEKFEKFGM-SPSKGVLFYGPPGCGKTLLAKAIANE 1708
Query: 410 AGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHE- 468
ANFIS+ G L + WFG++E + +F A AP ++F DE+DS+ RG +
Sbjct: 1709 CQANFISIKGPELLTMWFGESEANVREIFDKARGSAPCVLFFDELDSIATQRGSSVGDAG 1888
Query: 469 -ATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDADN 525
A R+ N+ + DG+ +K+ + I+GATNRP +D A++R RL + IY+ LPD D+
Sbjct: 1889 GAADRVLNQLLTEMDGMSAKKT--VFIIGATNRPDIIDPALLRPGRLDQLIYIPLPDEDS 2062
Query: 526 RKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE 579
R +I + L+ + D LA T G+SG+D+ +C A ++E +E++
Sbjct: 2063 RHQIFKACLRKSPISKDVDIRALAKYTQGFSGADITEICQRACKYAIRENIEKD 2224
Score = 166 bits (419), Expect = 3e-41
Identities = 101/285 (35%), Positives = 158/285 (55%), Gaps = 4/285 (1%)
Frame = +2
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V +DD+G + + ELV LP+R P+LF + +P KGILL+GPPG+GKTL+A+A+A
Sbjct: 707 VGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGV-KPPKGILLYGPPGSGKTLIARAVA 883
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E GA F ++G + SK G++E + F A K AP IIF+DE+DS+ R
Sbjct: 884 NETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-HG 1060
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDADN 525
E RR+ ++ + DGL+S+ + ++++GATNRP +D A+ R R R I + +PD
Sbjct: 1061EVERRIVSQLLTLMDGLKSRAH--VIVMGATNRPNSIDPALRRFGRFDREIDIGVPDEVG 1234
Query: 526 RKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNN 585
R ++LRI K+ L D +K++ T GY G+DL LC AA + ++E ++ D
Sbjct: 1235RLEVLRIHTKNMKLAEDVDLEKISKETHGYVGADLAALCTEAALQCIREKMDVIDLEDET 1414
Query: 586 ITNVALRPLNL--DDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
I L + + + F + PS + V W+++ G
Sbjct: 1415IDAEILNSMAVTNEHFATALGSSNPSALRETVVEVPNCSWDDIGG 1549
>AW696218 homologue to PIR|B96835|B96 CAD ATPase (AAA1) 35570-33019
[imported] - Arabidopsis thaliana, partial (34%)
Length = 556
Score = 166 bits (419), Expect = 3e-41
Identities = 80/174 (45%), Positives = 119/174 (67%), Gaps = 6/174 (3%)
Frame = +3
Query: 347 GVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKAL 406
GV++DD+ L + K+ L E V+LP+ PE F + RP KG+L+FGPPGTGKTL+AKA+
Sbjct: 39 GVRWDDVAGLTEAKRLLEEAVVLPLWMPEYFQ--GIRRPWKGVLMFGPPGTGKTLLAKAV 212
Query: 407 ATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFE 466
ATE G F +VS +TL SKW G++E++ + LF A AP IF+DE+DSL ARG + E
Sbjct: 213 ATECGTTFFNVSSATLASKWRGESERMVRCLFDLARAYAPSTIFIDEIDSLCNARGASGE 392
Query: 467 HEATRRMRNEFMAAWDGLRSK------ENQRILILGATNRPFDLDDAVIRRLPR 514
HE++RR+++E + DG+ + + +++L ATN P+D+D+A+ RRL +
Sbjct: 393 HESSRRVKSELLVQVDGVSNSATNEDGSRKLVMVLAATNFPWDIDEALRRRLEK 554
>TC87818 similar to GP|20197082|gb|AAC26698.2 putative katanin {Arabidopsis
thaliana}, partial (62%)
Length = 984
Score = 151 bits (381), Expect = 9e-37
Identities = 75/186 (40%), Positives = 121/186 (64%), Gaps = 1/186 (0%)
Frame = +2
Query: 394 PPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDE 453
P G + ++AKA+ATE F ++S S++ SKW GD+EKL K LF A AP IF+DE
Sbjct: 5 PQGLERPMLAKAVATECNTTFFNISASSMFSKWRGDSEKLVKVLFELARHHAPATIFLDE 184
Query: 454 VDSLLGARG-GAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRL 512
+D+++ RG G EHEA+RR++ E + DGL ++ ++ + +L ATN P++LD A++RRL
Sbjct: 185 IDAIISQRGEGRSEHEASRRLKTELLIQMDGL-ARTDELVFVLAATNLPWELDAAMLRRL 361
Query: 513 PRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPV 572
+RI V LP+ + R+ + +L + + +D L + T+GYSGSD++ LC A +P+
Sbjct: 362 EKRILVPLPEPEARRAMFEELLPLQPDEEPMPYDLLVDRTEGYSGSDIRLLCKETAMQPL 541
Query: 573 QELLEE 578
+ L+ +
Sbjct: 542 RRLMTQ 559
>TC77012 homologue to SP|P54776|PRSA_LYCES 26S protease regulatory subunit
6A homolog (TAT-binding protein homolog 1) (TBP-1)
(Mg(2+), complete
Length = 1793
Score = 150 bits (378), Expect = 2e-36
Identities = 119/388 (30%), Positives = 197/388 (50%), Gaps = 14/388 (3%)
Frame = +1
Query: 194 TSDDDEINKLFSNVVSIQPPKDENL---LATFKKQLEEDR-KIVISRSNLNELRKVLEEH 249
T D ++L N + I + + L ++K++++E++ KI +++ + ++E
Sbjct: 193 TDDIVRASRLLDNEIRILKEESQRTNLELESYKEKIKENQEKIKLNKQLPYLVGNIVEIL 372
Query: 250 QLSCVDLLHLDSDDVILTKQKAEKVVGWAKNHYLSSCLLPCV---KGDKLCLPRESLEIA 306
+++ D D ++ L Q+ K V K + LP V DKL P + + +
Sbjct: 373 EMNPEDEAEEDGANIDLDSQRKGKCVV-LKTSTRQTIFLPVVGLVDPDKL-KPGDLVGVN 546
Query: 307 ISRLKGVETMSRQPSQSLKNLAKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNEL 366
++T+ + +K + DE P E ++DIG LE + L E
Sbjct: 547 KDSYLILDTLPSEYDSRVKAMEVDE----------KPTE---DYNDIGGLEKQIQELVEA 687
Query: 367 VILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKW 426
++LPM E F + RP KG+LL+GPPGTGKTLMA+A A + A F+ ++G L +
Sbjct: 688 IVLPMTHKERFQKLGI-RPPKGVLLYGPPGTGKTLMARACAAQTNATFLKLAGPQLVQMF 864
Query: 427 FGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEAT-----RRMRNEFMAAW 481
GD KL + F A + +P IIF+DE+D++ R F+ E + +R E +
Sbjct: 865 IGDGAKLVRDAFQLAKEKSPCIIFIDEIDAIGTKR---FDSEVSGDREVQRTMLELLNQL 1035
Query: 482 DGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDADNRKKILRIILKHENL 539
DG S + RI ++ ATNR LD A++R RL R+I P + R +IL+I + N+
Sbjct: 1036DGFSS--DDRIKVIAATNRADILDPALMRSGRLDRKIEFPHPTEEARARILQIHSRKMNV 1209
Query: 540 DPDFQFDKLANLTDGYSGSDLKNLCITA 567
PD F++LA TD ++G+ LK +C+ A
Sbjct: 1210HPDVNFEELARSTDDFNGAQLKAVCVEA 1293
>TC76904 homologue to GP|4325041|gb|AAD17230.1| FtsH-like protein Pftf
precursor {Nicotiana tabacum}, partial (91%)
Length = 2667
Score = 147 bits (370), Expect = 2e-35
Identities = 96/256 (37%), Positives = 139/256 (53%), Gaps = 7/256 (2%)
Frame = +1
Query: 320 PSQSLKNLAKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTH 379
P Q ++ AK + E N GV FDD+ +++ K+ E+V +++PE FT
Sbjct: 766 PMQFGQSKAKFQMEPN----------TGVTFDDVAGVDEAKQDFMEVVEF-LKKPERFTS 912
Query: 380 GNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFS 439
P KG+LL GPPGTGKTL+AKA+A EAG F S+SGS + G + LF
Sbjct: 913 VGARIP-KGVLLVGPPGTGKTLLAKAIAGEAGVPFFSISGSEFVEMFVGIGASRVRDLFK 1089
Query: 440 FASKLAPVIIFVDEVDSL-----LGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILI 494
A + AP I+FVDE+D++ G GG E E T N+ + DG N +++
Sbjct: 1090 KAKENAPCIVFVDEIDAVGRQRGTGIGGGNDEREQT---LNQLLTEMDGFEG--NTGVIV 1254
Query: 495 LGATNRPFDLDDAVIR--RLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLT 552
+ ATNR LD A++R R R++ VD+PD R +IL++ ++ D D + +A T
Sbjct: 1255 VAATNRADILDSALLRPGRFDRQVSVDVPDVRGRTEILKVHANNKKFDNDVSLEVIAMRT 1434
Query: 553 DGYSGSDLKNLCITAA 568
G+SG+DL NL AA
Sbjct: 1435 PGFSGADLANLLNEAA 1482
>TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AAA-ATPase
subunit RPT4a-like protein {Arabidopsis thaliana},
complete
Length = 1640
Score = 146 bits (369), Expect = 2e-35
Identities = 87/240 (36%), Positives = 142/240 (58%), Gaps = 9/240 (3%)
Frame = +2
Query: 343 PGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLM 402
PG I + +G L D + L E + LP+ PELF + +P KG+LL+GPPGTGKTL+
Sbjct: 467 PGNIS--YSAVGGLSDQIRELRESIELPLMNPELFLRVGI-KPPKGVLLYGPPGTGKTLL 637
Query: 403 AKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGAR- 461
A+A+A+ ANF+ V S + K+ G++ +L + +F +A P IIF+DE+D++ G R
Sbjct: 638 ARAIASNIDANFLKVVSSAIIDKYIGESARLIREMFGYARDHQPCIIFMDEIDAIGGRRF 817
Query: 462 --GGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIY 517
G + + E R + E + DG + ++ ++ ATNRP LD A++R RL R+I
Sbjct: 818 SEGTSADREIQRTLM-ELLNQLDGF--DQLGKVKMIMATNRPDVLDPALLRPGRLDRKIE 988
Query: 518 VDLPDADNRKKILRI----ILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQ 573
+ LP+ +R +IL+I I KH +D ++ + L +G++G+DL+N+C A ++
Sbjct: 989 IPLPNEQSRMEILKIHAAGIAKHGEID----YEAVVKLAEGFNGADLRNVCTEAGMSAIR 1156
>TC77340 SP|Q9BAE0|FTSH_MEDSA Cell division protein ftsH homolog chloroplast
precursor (EC 3.4.24.-). [Alfalfa] {Medicago sativa},
complete
Length = 2473
Score = 144 bits (363), Expect = 1e-34
Identities = 95/248 (38%), Positives = 136/248 (54%), Gaps = 11/248 (4%)
Frame = +3
Query: 345 EIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAK 404
E GV F D+ + K L E+V ++ P+ +T P KG LL GPPGTGKTL+A+
Sbjct: 990 ETGVTFADVAGADQAKLELQEVVDF-LKNPDKYTALGAKIP-KGCLLVGPPGTGKTLLAR 1163
Query: 405 ALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSL-----LG 459
A+A EAG F S + S + G + LF A AP I+F+DE+D++ G
Sbjct: 1164 AVAGEAGTPFFSCAASEFVELFVGVGASRVRDLFEKAKSKAPCIVFIDEIDAVGRQRGAG 1343
Query: 460 ARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIY 517
GG E E T N+ + DG N +++L ATNRP LD A++R R R++
Sbjct: 1344 LGGGNDEREQT---INQLLTEMDGFSG--NSGVIVLAATNRPDVLDSALLRPGRFDRQVT 1508
Query: 518 VDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNL----CITAAYRPVQ 573
VD PD R KIL++ + + L D FDK+A T G++G+DL+NL I AA R ++
Sbjct: 1509 VDRPDVAGRVKILQVHSRGKALAKDVDFDKIARRTPGFTGADLQNLMNEAAILAARRDLK 1688
Query: 574 ELLEEEKA 581
E+ ++E A
Sbjct: 1689 EISKDEIA 1712
>TC85936 homologue to GP|13537115|dbj|BAB40755. AtSUG1 {Arabidopsis
thaliana}, partial (98%)
Length = 1633
Score = 144 bits (362), Expect = 1e-34
Identities = 87/230 (37%), Positives = 133/230 (57%), Gaps = 5/230 (2%)
Frame = +1
Query: 350 FDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATE 409
+D IG L+ K + E++ LP++ PELF + +P KG+LL+GPPGTGKTL+A+A+A
Sbjct: 619 YDMIGGLDQQIKEIKEVIELPIKHPELFESLGIAQP-KGVLLYGPPGTGKTLLARAVAHH 795
Query: 410 AGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGAR---GGAFE 466
FI VSGS L K+ G+ ++ + LF A + AP IIF+DE+DS+ AR G
Sbjct: 796 TDCTFIRVSGSELVQKYIGEGSRMVRELFVMAREHAPSIIFMDEIDSIGSARMESGSGNG 975
Query: 467 HEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDAD 524
+R E + DG + + +I +L ATNR LD A++R R+ R+I P+ +
Sbjct: 976 DSEVQRTMLELLNQLDGFEA--SNKIKVLMATNRIDILDQALLRPGRIDRKIEFPNPNEE 1149
Query: 525 NRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQE 574
+R+ IL+I + NL K+A +G SG++LK +C A ++E
Sbjct: 1150SRRDILKIHSRRMNLMRGIDLKKIAEKMNGASGAELKAVCTEAGMFALRE 1299
>TC85745 homologue to GP|17297987|dbj|BAB78491. 26S proteasome regulatory
particle triple-A ATPase subunit2b {Oryza sativa
(japonica cultivar-group), partial (98%)
Length = 1696
Score = 140 bits (354), Expect = 1e-33
Identities = 85/230 (36%), Positives = 129/230 (55%), Gaps = 5/230 (2%)
Frame = +3
Query: 350 FDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATE 409
+ DIG L+ + + E V LP+ PEL+ + +P KG++L+G PGTGKTL+AKA+A
Sbjct: 648 YADIGGLDAQIQEIKEAVELPLTHPELYEDIGI-KPPKGVILYGEPGTGKTLLAKAVANS 824
Query: 410 AGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAF---E 466
A F+ V GS L K+ GD KL + LF A L+P I+F+DE+D++ R A E
Sbjct: 825 TSATFLRVVGSELIQKYLGDGPKLVRELFRVADDLSPAIVFIDEIDAVGTKRYDAHSGGE 1004
Query: 467 HEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDAD 524
E R M E + DG S+ + ++++ ATNR LD A++R R+ R+I LPD
Sbjct: 1005REIQRTML-ELLNQLDGFDSRGDVKVIL--ATNRIESLDPALLRPGRIDRKIEFPLPDIK 1175
Query: 525 NRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQE 574
R++I +I L D ++ D +SG+D+K +C A ++E
Sbjct: 1176TRRRIFQIHTSRMTLADDVNLEEFVMTKDEFSGADIKAICTEAGLLALRE 1325
>TC85744 homologue to PIR|T08959|T08959 proteinase homolog F19B15.70 -
Arabidopsis thaliana, complete
Length = 1629
Score = 139 bits (350), Expect = 3e-33
Identities = 85/230 (36%), Positives = 128/230 (54%), Gaps = 5/230 (2%)
Frame = +2
Query: 350 FDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATE 409
+ DIG L+ + + E V LP+ PEL+ + +P KG++L+G PGTGKTL+AKA+A
Sbjct: 650 YADIGGLDAQIQEIKEAVELPLTHPELYEDIGI-KPPKGVILYGEPGTGKTLLAKAVANS 826
Query: 410 AGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAF---E 466
A F+ V GS L K+ GD KL + LF A L+P I+F+DE+D++ R A E
Sbjct: 827 TSATFLRVVGSELIQKYLGDGPKLVRELFRVADDLSPSIVFIDEIDAVGTKRYDAHSGGE 1006
Query: 467 HEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDAD 524
E R M E + DG S+ + ++++ ATNR LD A++R R+ R+I LPD
Sbjct: 1007REIQRTML-ELLNQLDGFDSRGDVKVIL--ATNRIESLDPALLRPGRIDRKIEFPLPDIK 1177
Query: 525 NRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQE 574
R++I I L D ++ D +SG+D+K +C A ++E
Sbjct: 1178TRRRIFTIHTSRMTLADDVNLEEFVMTKDEFSGADIKAICTEAGLLALRE 1327
>TC84403 homologue to GP|15021761|gb|AAK77908.1 AAA-metalloprotease FtsH
{Pisum sativum}, partial (36%)
Length = 897
Score = 134 bits (337), Expect = 1e-31
Identities = 78/189 (41%), Positives = 111/189 (58%), Gaps = 7/189 (3%)
Frame = +3
Query: 387 KGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAP 446
KG LL GPPGTGKTL+AKA A E+G F+S+SGS + G + LF A K AP
Sbjct: 18 KGALLVGPPGTGKTLLAKATAGESGVPFLSISGSDFLELFVGVGSSRVRNLFKEARKCAP 197
Query: 447 VIIFVDEVDSL---LGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFD 503
I+F+DE+D++ G+RGG ++ N+ + DG + +++L TNRP
Sbjct: 198 SIVFIDEIDAIGRARGSRGGDRANDERESTLNQLLVEMDGFGTTAG--VVVLAGTNRPDV 371
Query: 504 LDDAVIR--RLPRRIYVDLPDADNRKKILRIILKHENLD--PDFQFDKLANLTDGYSGSD 559
LD A++R R R+I +D PD R +I +I LK LD P + KLA+LT G++G+D
Sbjct: 372 LDKALLRPGRFDRQITIDKPDIKGRDQIFQIYLKGIKLDHEPSYYSHKLASLTPGFAGAD 551
Query: 560 LKNLCITAA 568
+ N+C AA
Sbjct: 552 IANVCNEAA 578
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,996,324
Number of Sequences: 36976
Number of extensions: 225184
Number of successful extensions: 1219
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 1152
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1168
length of query: 645
length of database: 9,014,727
effective HSP length: 102
effective length of query: 543
effective length of database: 5,243,175
effective search space: 2847044025
effective search space used: 2847044025
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0135.9


