

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0135.8
(1406 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
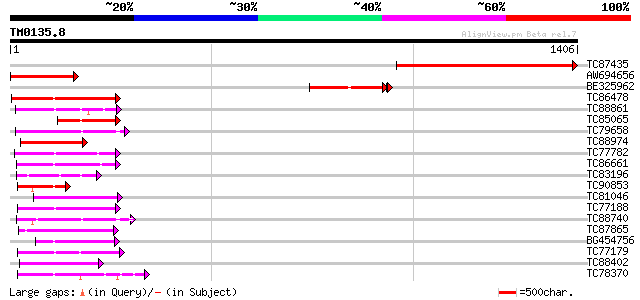
Score E
Sequences producing significant alignments: (bits) Value
TC87435 similar to GP|3549652|emb|CAA12272.1 MAP3K epsilon prote... 842 0.0
AW694656 homologue to GP|9280305|db MAP3K epsilon protein kinase... 303 4e-82
BE325962 similar to GP|3549652|emb MAP3K epsilon protein kinase ... 280 1e-77
TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 prote... 204 1e-52
TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative pro... 195 9e-50
TC85065 similar to GP|1255448|dbj|BAA09057.1 mitogen-activated p... 142 7e-34
TC79658 similar to PIR|S29851|S29851 protein kinase 6 (EC 2.7.1.... 140 5e-33
TC88974 similar to GP|3819699|emb|CAA08758.1 BnMAP4K alpha2 {Bra... 133 4e-31
TC77782 similar to GP|13448037|gb|AAK26845.1 SOS2-like protein k... 129 1e-29
TC86661 similar to GP|9798599|dbj|BAB11737.1 serine/threonine pr... 128 2e-29
TC83196 similar to GP|7270830|emb|CAB80511.1 protein kinase like... 126 5e-29
TC90853 similar to GP|2342427|dbj|BAA21857.1 NPK1-related protei... 126 7e-29
TC81046 similar to GP|17529280|gb|AAL38867.1 putative protein ki... 124 3e-28
TC77188 similar to PIR|T04862|T04862 probable serine/threonine-s... 124 3e-28
TC88740 similar to PIR|S29851|S29851 protein kinase 6 (EC 2.7.1.... 124 3e-28
TC87865 homologue to GP|13936371|gb|AAK40361.1 CTR1-like protein... 122 7e-28
BG454756 homologue to GP|12718822|db NQK1 MAPKK {Nicotiana tabac... 122 1e-27
TC77179 similar to GP|2665890|gb|AAB88537.1| calcium-dependent p... 121 2e-27
TC88402 similar to GP|20804521|dbj|BAB92215. putative CRK1 prote... 120 3e-27
TC78370 similar to GP|5001828|gb|AAD37165.1| 3-phosphoinositide-... 120 4e-27
>TC87435 similar to GP|3549652|emb|CAA12272.1 MAP3K epsilon protein kinase
{Arabidopsis thaliana}, partial (29%)
Length = 1646
Score = 842 bits (2176), Expect = 0.0
Identities = 430/448 (95%), Positives = 441/448 (97%)
Frame = +3
Query: 959 SGRFSGQLEYVRQFSGLERHESVLPLLHATEKKTNGELDFLMAEFADVSQRGRENGNLDS 1018
SGRFSGQLEYVRQFS LERHESVLPLLHA+E KTNGELDFLMAEFADVSQRGRENGNLDS
Sbjct: 6 SGRFSGQLEYVRQFSALERHESVLPLLHASENKTNGELDFLMAEFADVSQRGRENGNLDS 185
Query: 1019 SARVSHKVAPKKLGTFGSSEGAASTSGIVSQTASGVLSGSGVLNARPGSATSSGLLSHMV 1078
SARVS +VAPKKLGTFGSSEGAASTSGIVSQTASGVLSGSGVLNARP SATSSGLLSHMV
Sbjct: 186 SARVSQRVAPKKLGTFGSSEGAASTSGIVSQTASGVLSGSGVLNARPCSATSSGLLSHMV 365
Query: 1079 SSLNADVAREYLEKVADLLLEFAQADTTVKSYMCSQSLLSRLFQMFNRVEPPILLKILKC 1138
SSLNA+VA+EYLEKVADLLLEFAQADTTVKSYMCSQ+LLSRLFQMFNRVEPPILLKIL+C
Sbjct: 366 SSLNAEVAKEYLEKVADLLLEFAQADTTVKSYMCSQTLLSRLFQMFNRVEPPILLKILRC 545
Query: 1139 INHLSTDPNCLENLQRAEAIKHLIPNLELKEGTLVSEIHHEVLNALFNLCKINKRRQEQA 1198
INHLSTDPNCLENLQRAEAIK+LIPNLELKEG+LVSEIHHEVLNALFNLCKINKRRQEQA
Sbjct: 546 INHLSTDPNCLENLQRAEAIKYLIPNLELKEGSLVSEIHHEVLNALFNLCKINKRRQEQA 725
Query: 1199 AENGIIPHLMQFITSNSPLKQYALPLLCDMAHASRNSREQLRAHGGLDVYLNLLEDELWS 1258
AENGIIPHLMQFITSNSPLKQYALPLLCDMAHASRNSREQLRAHGGLDVYLNLLEDE WS
Sbjct: 726 AENGIIPHLMQFITSNSPLKQYALPLLCDMAHASRNSREQLRAHGGLDVYLNLLEDEFWS 905
Query: 1259 VTALDSIAVCLAHDNDNRKVEQSLLKKDAVQKLVKFFQSCPEQHFVHILEPFLKIITKSS 1318
VTALDSIAVCLAHDNDNRKVEQ+LLKKDAVQKLV FFQSCPE HFVHILEPFLKIITKS+
Sbjct: 906 VTALDSIAVCLAHDNDNRKVEQALLKKDAVQKLVMFFQSCPEPHFVHILEPFLKIITKSA 1085
Query: 1319 RINTTLAVNGLTPLLIARLDHQDAIARLNLLRLIKAVYEHHPQPKKLIVENDLPEKLQNL 1378
RINTTLAVNGLTPLL+ARLDHQDAIARLNLLRLIKAVYEHHPQPKKLIVENDLPEKLQNL
Sbjct: 1086 RINTTLAVNGLTPLLVARLDHQDAIARLNLLRLIKAVYEHHPQPKKLIVENDLPEKLQNL 1265
Query: 1379 IGERRDGQVLVKQMATSLLKALHINTVL 1406
IGERRDGQVLVKQMATSLLKALHINTVL
Sbjct: 1266 IGERRDGQVLVKQMATSLLKALHINTVL 1349
>AW694656 homologue to GP|9280305|db MAP3K epsilon protein kinase
{Arabidopsis thaliana}, partial (12%)
Length = 651
Score = 303 bits (775), Expect = 4e-82
Identities = 153/169 (90%), Positives = 160/169 (94%)
Frame = +2
Query: 1 MSRQSTSSAFTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQED 60
MSRQST SAFT+SKTLDNKYMLGDEIGKGAY RVYKGLDLENGDFVAIKQVSLENIAQED
Sbjct: 122 MSRQSTGSAFTQSKTLDNKYMLGDEIGKGAYARVYKGLDLENGDFVAIKQVSLENIAQED 301
Query: 61 LNIIMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESL 120
LNIIM+ I++L+NLNHKNIVKYLGSLKTKSHLHI+LEYVENGSLANIIKPNKFGPFPESL
Sbjct: 302 LNIIMQEIDLLKNLNHKNIVKYLGSLKTKSHLHIVLEYVENGSLANIIKPNKFGPFPESL 481
Query: 121 VAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKL 169
VAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKL F K+
Sbjct: 482 VAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLC*FWCCNKI 628
>BE325962 similar to GP|3549652|emb MAP3K epsilon protein kinase {Arabidopsis
thaliana}, partial (4%)
Length = 599
Score = 280 bits (717), Expect(2) = 1e-77
Identities = 149/193 (77%), Positives = 157/193 (81%)
Frame = +1
Query: 743 LQQTTPRNDFCRIAAKNGILLRLINTLYSLNESTRLASMSAGGGFLADGSAQRPRSGILD 802
LQQ+TPRNDFCRIAAKNGILLRLINTLYSLNESTRLASMSAG GFL DGS QRPRSGILD
Sbjct: 1 LQQSTPRNDFCRIAAKNGILLRLINTLYSLNESTRLASMSAGSGFLVDGSTQRPRSGILD 180
Query: 803 PAHPFMNQNDAQLSSADQQDLSKVRRGVLDHHLEPMHASSSNPRRSDANYPTDVDRPQSS 862
P HPF QN+A LSSADQQDL+K+R G LDHHLE H RRSD+NY DVDRPQSS
Sbjct: 181 PTHPFFGQNEALLSSADQQDLTKLRHGALDHHLESSH------RRSDSNYQMDVDRPQSS 342
Query: 863 NAAAEAVSLGKSLNLTSRESSVVALKERENVDRWKTDPSRAEVEPRQQRSSISANRTSTD 922
NAAAEAV L SLNL SRESS LKEREN DRWK+DPSRA+VE R QR SIS NR STD
Sbjct: 343 NAAAEAVPLEMSLNLASRESSAGTLKERENADRWKSDPSRADVELR-QRLSISGNRKSTD 519
Query: 923 RPPKLAEPSSNGL 935
R KL E SSNG+
Sbjct: 520 RSSKLTETSSNGV 558
Score = 30.0 bits (66), Expect(2) = 1e-77
Identities = 13/15 (86%), Positives = 14/15 (92%)
Frame = +2
Query: 934 GLSMTGATQQEQVRP 948
GLS TGATQQEQV+P
Sbjct: 554 GLSATGATQQEQVKP 598
>TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 protein kinase
{Brassica napus}, partial (52%)
Length = 2266
Score = 204 bits (520), Expect = 1e-52
Identities = 111/275 (40%), Positives = 167/275 (60%), Gaps = 5/275 (1%)
Frame = +2
Query: 5 STSSAFTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSL---ENIAQEDL 61
+T S F S +K+ G +G+G +G VY G + ENG AIK+V + + ++E L
Sbjct: 926 NTRSPFENSSPNLSKWKKGKLLGRGTFGHVYLGFNSENGQMCAIKEVRVGCDDQNSKECL 1105
Query: 62 NIIMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLV 121
+ + I++L L+H NIV+YLGS + L + LEYV GS+ +++ ++GPF E ++
Sbjct: 1106 KQLHQEIDLLNQLSHPNIVQYLGSELGEESLSVYLEYVSGGSIHKLLQ--EYGPFKEPVI 1279
Query: 122 AVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVG 181
Y Q++ GL YLH + +HRDIKGANIL G +KLADFG+A +T A + S G
Sbjct: 1280 QNYTRQIVSGLAYLHGRNTVHRDIKGANILVDPNGEIKLADFGMAKHITSA-ASMLSFKG 1456
Query: 182 TPYWMAPEVI-EMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQD-EQPPI 239
+PYWMAPEV+ +G DIWS+GCT+IE+ PP+ + + A+F+I + P I
Sbjct: 1457 SPYWMAPEVVMNTNGYSLPVDIWSLGCTLIEMAASKPPWSQYEGVAAIFKIGNSKDMPII 1636
Query: 240 PDSLSPNITDFLHQCFKKDARQRPDAKTLLSHPWI 274
P+ LS + +F+ C ++D RP A+ LL HP+I
Sbjct: 1637 PEHLSNDAKNFIMLCLQRDPSARPTAQKLLEHPFI 1741
>TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative protein kinase
{Oryza sativa (japonica cultivar-group)}, partial (34%)
Length = 1847
Score = 195 bits (496), Expect = 9e-50
Identities = 112/272 (41%), Positives = 158/272 (57%), Gaps = 10/272 (3%)
Frame = +1
Query: 15 TLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEM---INIL 71
++ + G IG+G++G VY +LE G A+K+V L + + I ++ I IL
Sbjct: 1009 SMKGHWQKGKLIGRGSFGSVYHATNLETGASCALKEVDLVPDDPKSTDCIKQLDQEIRIL 1188
Query: 72 QNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEG 131
L+H NIV+Y GS L I +EYV GSL ++ + G ES+V + +L G
Sbjct: 1189 GQLHHPNIVEYYGSEVVGDRLCIYMEYVHPGSLQKFMQDH-CGVMTESVVRNFTRHILSG 1365
Query: 132 LVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEVI 191
L YLH IHRDIKGAN+L G+VKLADFGV+ LTE S+ G+PYWMAPE++
Sbjct: 1366 LAYLHSTKTIHRDIKGANLLVDASGIVKLADFGVSKILTEKSYEL-SLKGSPYWMAPELM 1542
Query: 192 EMS-------GVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPIPDSLS 244
+ V A DIWS+GCT+IE+LT PP+ + A+F+++ P IP +LS
Sbjct: 1543 MAAMKNETNPTVAMAVDIWSLGCTIIEMLTGKPPWSEFPGHQAMFKVLH-RSPDIPKTLS 1719
Query: 245 PNITDFLHQCFKKDARQRPDAKTLLSHPWILN 276
P DFL QCF+++ RP A LL+HP++ N
Sbjct: 1720 PEGQDFLEQCFQRNPADRPSAAVLLTHPFVQN 1815
>TC85065 similar to GP|1255448|dbj|BAA09057.1 mitogen-activated protein
kinase {Arabidopsis thaliana}, partial (26%)
Length = 789
Score = 142 bits (359), Expect = 7e-34
Identities = 72/159 (45%), Positives = 104/159 (65%), Gaps = 2/159 (1%)
Frame = +2
Query: 118 ESLVAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTH 177
+S V+ Y Q+L GL YLH++ ++HRDIK ANIL G VK+ADFG++ + D+
Sbjct: 17 DSQVSAYTRQILHGLKYLHDRNIVHRDIKCANILVDANGSVKVADFGLS*AIKLNDVK-- 190
Query: 178 SVVGTPYWMAPEVI--EMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDE 235
S GT +WMAPEV+ ++ G +DIWS+GCTV+E+LT PY ++ + A+FRI + E
Sbjct: 191 SCQGTAFWMAPEVVRGKVKGYGLPADIWSLGCTVLEMLTGQVPYAPMECISAMFRIGKGE 370
Query: 236 QPPIPDSLSPNITDFLHQCFKKDARQRPDAKTLLSHPWI 274
PP+PD+LS + DF+ QC K + RP A LL H ++
Sbjct: 371 LPPVPDTLSRDARDFILQCLKVNPDDRPTAAQLLDHKFV 487
>TC79658 similar to PIR|S29851|S29851 protein kinase 6 (EC 2.7.1.-) -
soybean, partial (78%)
Length = 1517
Score = 140 bits (352), Expect = 5e-33
Identities = 88/289 (30%), Positives = 147/289 (50%), Gaps = 7/289 (2%)
Frame = +2
Query: 15 TLD-NKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLEN----IAQEDLNIIMEMIN 69
T+D +K LG + GA+ R+Y G+ E V I +V ++ +A + N + +
Sbjct: 299 TIDMSKLFLGHKFAHGAHSRLYHGVYKEESVAVKIIKVPDDDENGELASKLENQFVREVT 478
Query: 70 ILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVL 129
+L L+H+N++K++ + + II EY+ GSL + + P + + +
Sbjct: 479 LLSRLHHRNVIKFIAASRNPPVYCIITEYLSEGSLRAYLHKLEHKAIPLQKLIAFALDIS 658
Query: 130 EGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPE 189
G+ Y+H QGVIHRD+K N+L ++ +KLADFG+A + D+ GT WMAPE
Sbjct: 659 RGMAYIHSQGVIHRDLKPENVLIDEDFRLKLADFGIACEEAVCDLLADD-PGTYRWMAPE 835
Query: 190 VIEMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDE-QPPIPDSLSPNIT 248
+I+ D++S G + E+LT PY D+ P+ A F +V + +P IP + P +
Sbjct: 836 MIKRKSYGRKVDVYSFGLILWEMLTGTIPYEDMNPIQAAFAVVNKKLRPVIPSNCPPAMR 1015
Query: 249 DFLHQCFKKDARQRPDAKTLLSHPW-ILNCRRVLQSSLRHSGTLRTIKD 296
+ QC+ +RPD W I+ +SSL GTL +++
Sbjct: 1016ALIEQCWSLQPDKRPDF-------WQIVKVLEQFESSLARDGTLTLVQN 1141
>TC88974 similar to GP|3819699|emb|CAA08758.1 BnMAP4K alpha2 {Brassica
napus}, partial (34%)
Length = 851
Score = 133 bits (335), Expect = 4e-31
Identities = 71/167 (42%), Positives = 106/167 (62%)
Frame = +3
Query: 26 IGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNIVKYLGS 85
IG+G++G VYKG D E VAIK + LE +++D++ I + I++L I +Y GS
Sbjct: 207 IGQGSFGDVYKGFDKELNKEVAIKVIDLEE-SEDDIDDIQKEISVLSQCRCPYITEYYGS 383
Query: 86 LKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQGVIHRDI 145
++ L II+EY+ GS+A++++ P E +A + +L + YLH +G IHRDI
Sbjct: 384 FLNQTKLWIIMEYMAGGSVADLLQSGP--PLDEMSIAYILRDLLHAVDYLHNEGKIHRDI 557
Query: 146 KGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEVIE 192
K ANIL ++ G VK+ADFGV+ +LT + VGTP+WMAPEVI+
Sbjct: 558 KAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWMAPEVIQ 698
>TC77782 similar to GP|13448037|gb|AAK26845.1 SOS2-like protein kinase PKS6
{Arabidopsis thaliana}, partial (77%)
Length = 1236
Score = 129 bits (323), Expect = 1e-29
Identities = 78/264 (29%), Positives = 137/264 (51%), Gaps = 2/264 (0%)
Frame = +2
Query: 12 KSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDL-NIIMEMINI 70
+ +T KY +G IG+G++ +V ++ENGD+VAIK + ++ + ++ + + I+
Sbjct: 167 RPRTRVGKYEMGKTIGEGSFAKVKLAKNVENGDYVAIKILDRNHVLKHNMMDQLKREISA 346
Query: 71 LQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLE 130
++ +NH N++K + +K+ ++I+LE V G L + I + G E Y Q++
Sbjct: 347 MKIINHPNVIKIFEVMASKTKIYIVLELVNGGELFDKIATH--GRLKEDEARSYFQQLIN 520
Query: 131 GLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEV 190
+ Y H +GV HRD+K N+L G++K++DFG++T + D + GTP ++APEV
Sbjct: 521 AVDYCHSRGVYHRDLKPENLLLDTNGVLKVSDFGLSTYSQQEDELLRTACGTPNYVAPEV 700
Query: 191 IEMSG-VCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPIPDSLSPNITD 249
+ G V ++SDIWS G + L+ P +D + AL+R + P SP
Sbjct: 701 LNDRGYVGSSSDIWSCGVILFVLMAGYLP-FDEPNLIALYRKIGRADFKCPSWFSPEAKK 877
Query: 250 FLHQCFKKDARQRPDAKTLLSHPW 273
L + R +L W
Sbjct: 878 LLTSILNPNPLTRIKIPEILEDEW 949
>TC86661 similar to GP|9798599|dbj|BAB11737.1 serine/threonine protein
kinase {Arabidopsis thaliana}, partial (58%)
Length = 1840
Score = 128 bits (321), Expect = 2e-29
Identities = 83/262 (31%), Positives = 134/262 (50%), Gaps = 4/262 (1%)
Frame = +2
Query: 16 LDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDL-NIIMEMINILQNL 74
L KY LG +G GA+ +VY + ENG VA+K ++ + I+ L + I+I+ L
Sbjct: 293 LFGKYELGKLLGCGAFAKVYHARNTENGQSVAVKVINKKKISATGLAGHVKREISIMSKL 472
Query: 75 NHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVY 134
H NIV+ L TK+ ++ ++E+ + G L I NK G F E L Q++ + Y
Sbjct: 473 RHPNIVRLHEVLATKTKIYFVMEFAKGGELFAKIA-NK-GRFSEDLSRRLFQQLISAVGY 646
Query: 135 LHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADIN--THSVVGTPYWMAPEVIE 192
H GV HRD+K N+L +G +K++DFG++ + I+ H++ GTP ++APE++
Sbjct: 647 CHSHGVFHRDLKPENLLLDDKGNLKVSDFGLSAVKEQVRIDGMLHTLCGTPAYVAPEILA 826
Query: 193 MSGVCAAS-DIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPIPDSLSPNITDFL 251
G A DIWS G + L+ P+ D M ++R + + P SP++ FL
Sbjct: 827 KRGYDGAKVDIWSCGIILFVLVAGYLPFNDPNLM-VMYRKIYSGEFKCPRWFSPDLKRFL 1003
Query: 252 HQCFKKDARQRPDAKTLLSHPW 273
+ + R +L PW
Sbjct: 1004SRLMDTNPETRITVDEILRDPW 1069
>TC83196 similar to GP|7270830|emb|CAB80511.1 protein kinase like protein
{Arabidopsis thaliana}, partial (66%)
Length = 1484
Score = 126 bits (317), Expect = 5e-29
Identities = 78/215 (36%), Positives = 124/215 (57%), Gaps = 3/215 (1%)
Frame = +3
Query: 16 LDNKYM-LGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNL 74
+D K++ G +I +YG +YKG+ VAIK + E+++ E + + I++ +
Sbjct: 858 IDPKHLKYGTQIASASYGELYKGIYCSQE--VAIKVLKAEHVSSEMQKEFAQEVYIMRKV 1031
Query: 75 NHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNK-FGPFPESL-VAVYIAQVLEGL 132
HKN+V+++G+ L I+ E++ GS+ + + K F FP L VA+ V +G+
Sbjct: 1032 RHKNVVQFMGACTQPPRLCIVTEFMSGGSVYDYLHKQKGFFKFPTVLKVAI---DVSKGM 1202
Query: 133 VYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEVIE 192
YLH+ +IHRD+K AN+L + G+VK+ADFGVA ++ + T + GT WMAPEVIE
Sbjct: 1203 NYLHQHNIIHRDLKAANLLMDENGVVKVADFGVARVRAQSGVMT-AETGTYRWMAPEVIE 1379
Query: 193 MSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPA 227
+D++S G + ELLT PY L P+ A
Sbjct: 1380 HKPYDHKADVFSFGVVLWELLTGKLPYEFLTPLQA 1484
>TC90853 similar to GP|2342427|dbj|BAA21857.1 NPK1-related protein kinase 3
{Arabidopsis thaliana}, partial (22%)
Length = 746
Score = 126 bits (316), Expect = 7e-29
Identities = 61/139 (43%), Positives = 96/139 (68%), Gaps = 6/139 (4%)
Frame = +3
Query: 19 KYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLE------NIAQEDLNIIMEMINILQ 72
++ G+ IG GA+GRVY G++L++G+ +A+KQV +E + ++ + E + +L+
Sbjct: 258 RWRKGELIGSGAFGRVYMGMNLDSGELIAVKQVLIEPGIAFKENTKANIRELEEEVKLLK 437
Query: 73 NLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGL 132
NL H NIV+YLG+ + + L+I+LE+V GS+++++ KFG FPES + Y Q+L+GL
Sbjct: 438 NLKHPNIVRYLGTAREEDSLNILLEFVPGGSISSLL--GKFGSFPESGITTYTKQLLDGL 611
Query: 133 VYLHEQGVIHRDIKGANIL 151
YLH +IHRDIKGANIL
Sbjct: 612 EYLHNNRIIHRDIKGANIL 668
>TC81046 similar to GP|17529280|gb|AAL38867.1 putative protein kinase
{Arabidopsis thaliana}, partial (44%)
Length = 921
Score = 124 bits (311), Expect = 3e-28
Identities = 72/227 (31%), Positives = 120/227 (52%), Gaps = 8/227 (3%)
Frame = +2
Query: 60 DLNIIMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPES 119
+L+ I + + ++H N+++ S L +++ ++ GS +I+K + F E
Sbjct: 11 NLDGIRREVQTMSLIDHPNLLRAHCSFTAGHSLWVVMPFMSGGSCLHIMKSSFPEGFDEP 190
Query: 120 LVAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEA---DINT 176
++A + +VL+ LVYLH G IHRD+K NIL G VK+ADFGV+ + + +
Sbjct: 191 VIATVLREVLKALVYLHAHGHIHRDVKAGNILLDANGSVKMADFGVSACMFDTGDRQRSR 370
Query: 177 HSVVGTPYWMAPEVI-EMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDE 235
++ VGTP WMAPEV+ ++ G +DIWS G T +EL P+ PM L +Q+
Sbjct: 371 NTFVGTPCWMAPEVMQQLHGYDFKADIWSFGITALELAHGHAPFSKYPPMKVLLMTLQNA 550
Query: 236 QPPI----PDSLSPNITDFLHQCFKKDARQRPDAKTLLSHPWILNCR 278
P + S + + + C KD ++RP ++ LL H + + R
Sbjct: 551 PPGLDYERDKRFSKSFKELVATCLVKDPKKRPSSEKLLKHHFFKHAR 691
>TC77188 similar to PIR|T04862|T04862 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) F28A21.110 - Arabidopsis
thaliana, partial (79%)
Length = 1881
Score = 124 bits (311), Expect = 3e-28
Identities = 75/259 (28%), Positives = 134/259 (50%), Gaps = 4/259 (1%)
Frame = +1
Query: 19 KYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDL-NIIMEMINILQNLNHK 77
++ LG +G G + +V+ +L+ G+ VAIK +S + I + L + I I+IL+ + H
Sbjct: 187 RFELGKLLGHGTFAKVHLAKNLKTGESVAIKIISKDKILKSGLVSHIKREISILRRVRHP 366
Query: 78 NIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHE 137
NIV+ + TK+ ++ ++EYV G L N + G E + Y Q++ + + H
Sbjct: 367 NIVQLFEVMATKTKIYFVMEYVRGGELFNKVAK---GRLKEEVARKYFQQLICAVGFCHA 537
Query: 138 QGVIHRDIKGANILTTKEGLVKLADFGVATKLTE--ADINTHSVVGTPYWMAPEVIEMSG 195
+GV HRD+K N+L ++G +K++DFG++ E D H+ GTP ++APEV+ G
Sbjct: 538 RGVFHRDLKPENLLLDEKGNLKVSDFGLSAVSDEIKQDGLFHTFCGTPAYVAPEVLSRKG 717
Query: 196 VCAAS-DIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPIPDSLSPNITDFLHQC 254
A DIWS G + L+ P++D + A+++ + + P SP + L +
Sbjct: 718 YDGAKVDIWSCGVVLFVLMAGYLPFHDPNNVMAMYKKIYKGEFRCPRWFSPELVSLLTRL 897
Query: 255 FKKDARQRPDAKTLLSHPW 273
+ R ++ + W
Sbjct: 898 LDIKPQTRISIPEIMENRW 954
>TC88740 similar to PIR|S29851|S29851 protein kinase 6 (EC 2.7.1.-) -
soybean, partial (75%)
Length = 1364
Score = 124 bits (310), Expect = 3e-28
Identities = 86/300 (28%), Positives = 145/300 (47%), Gaps = 7/300 (2%)
Frame = +2
Query: 18 NKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLEN-----IAQEDLNIIMEMINILQ 72
+K +G GA+ R+Y G+ E V I +V ++ A + I E + +L
Sbjct: 197 SKLFVGLRFAYGAHSRLYHGMYEEEPVAVKIIRVPDDDENGTMAATLEKQFITE-VTLLS 373
Query: 73 NLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGL 132
L+H+N++K++ + + +I EY+ GSL + + + + + G+
Sbjct: 374 RLHHQNVLKFVAACRKPPVYCVITEYLLEGSLRAYLHKLEGKIISLQKLIAFSLDIARGM 553
Query: 133 VYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEVIE 192
Y+H QGVIHRD+K N+L + +K+ADFG+A + D+ GT WMAPE+I
Sbjct: 554 EYIHSQGVIHRDLKPENVLINGDSHLKIADFGIACEEAYCDLLADD-PGTYRWMAPEMIR 730
Query: 193 MSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIV-QDEQPPIPDSLSPNITDFL 251
D++S G + E+LT PY D+ P+ A F +V ++ +P IP + P + +
Sbjct: 731 RKSYGRKVDVYSFGLILWEMLTGTIPYEDMTPIQAAFAVVNKNSRPIIPSNCPPAMRALI 910
Query: 252 HQCFKKDARQRPDAKTLLSHPW-ILNCRRVLQSSLRHSGTLRTIKDDGSAVAEVSGGDHK 310
QC+ + +RPD W ++ +SSL GTL +++ G DHK
Sbjct: 911 EQCWSLNPDKRPDF-------WQVVKVLEQFESSLARDGTLTLLQNP-------CGQDHK 1048
>TC87865 homologue to GP|13936371|gb|AAK40361.1 CTR1-like protein kinase
{Rosa hybrid cultivar}, partial (31%)
Length = 1190
Score = 122 bits (307), Expect = 7e-28
Identities = 74/253 (29%), Positives = 133/253 (52%), Gaps = 4/253 (1%)
Frame = +3
Query: 21 MLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNIV 80
+L ++IG G++G V++ NG VA+K + ++ E M + I+++L H NIV
Sbjct: 60 VLKEKIGSGSFGTVHRAE--WNGSDVAVKILMEQDFHAERFKEFMREVAIMKHLRHPNIV 233
Query: 81 KYLGSLKTKSHLHIILEYVENGSLANII-KPNKFGPFPESLVAVYIAQVLEGLVYLHEQG 139
+G++ +L I+ EY+ GSL ++ +P E V +G+ YLH++
Sbjct: 234 LLMGAVTQPPNLSIVTEYLSRGSLYRLLHRPGAKEVLDERRRLSMAYDVAKGMNYLHKRN 413
Query: 140 --VIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEVIEMSGVC 197
++HRD+K N+L K+ VK+ DFG++ +++ S GTP WMAPEV+
Sbjct: 414 PPIVHRDLKSPNLLVDKKYTVKVCDFGLSRLKANTFLSSKSAAGTPEWMAPEVLRDEPSN 593
Query: 198 AASDIWSVGCTVIELLTCVPPYYDLQPMPALFRI-VQDEQPPIPDSLSPNITDFLHQCFK 256
SDI+S G + E+ T P+ +L P + + + ++ IP +L+P I + C+
Sbjct: 594 EKSDIYSFGVIMWEIATLQQPWGNLNPAQVVAAVGFKHKRLEIPRNLNPQIAAIIEACWA 773
Query: 257 KDARQRPDAKTLL 269
+ +RP +++
Sbjct: 774 NEPWKRPSFASIM 812
>BG454756 homologue to GP|12718822|db NQK1 MAPKK {Nicotiana tabacum}, partial
(60%)
Length = 648
Score = 122 bits (306), Expect = 1e-27
Identities = 80/217 (36%), Positives = 115/217 (52%), Gaps = 9/217 (4%)
Frame = +2
Query: 64 IMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAV 123
I++ + I Q ++V S + ++LEY++ GSL ++I+ + E +AV
Sbjct: 2 IVQELKINQASQCPHVVVCYHSFYNNGVISLVLEYMDRGSLVDVIR--QVNTILEPYLAV 175
Query: 124 YIAQVLEGLVYLH-EQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGT 182
QVL+GLVYLH E+ VIHRDIK +N+L +G VK+ DFGV+ L + VGT
Sbjct: 176 VCKQVLQGLVYLHNERHVIHRDIKPSNLLVNHKGEVKITDFGVSAMLASTMGQRDTFVGT 355
Query: 183 PYWMAPEVIEMSGVCAASDIWSVGCTVIELLTCVPPYY---DLQPMPALFRIVQD--EQP 237
+M+PE I S + DIWS+G V+E PY D Q P+ + ++Q E P
Sbjct: 356 YNYMSPERISGSTYDYSCDIWSLGMVVLECAIGRFPYIQSEDQQAWPSFYELLQAIVESP 535
Query: 238 P---IPDSLSPNITDFLHQCFKKDARQRPDAKTLLSH 271
P PD SP F+ C KKD R+R + LL H
Sbjct: 536 PPSAPPDQFSPEFCSFVSSCIKKDPRERSTSLELLDH 646
>TC77179 similar to GP|2665890|gb|AAB88537.1| calcium-dependent protein
kinase {Fragaria x ananassa}, partial (92%)
Length = 2243
Score = 121 bits (304), Expect = 2e-27
Identities = 80/275 (29%), Positives = 138/275 (50%), Gaps = 8/275 (2%)
Frame = +1
Query: 19 KYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQE-DLNIIMEMINILQNL-NH 76
+Y LG E+G+G +G Y D E G+ +A K +S + + D+ + + I+++L H
Sbjct: 391 QYELGRELGRGEFGITYLCKDRETGEELACKSISKDKLRTAIDIEDVRREVEIMRHLPKH 570
Query: 77 KNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLH 136
NIV + + ++H+++E E G L + I G + E A + +++ + H
Sbjct: 571 PNIVTLKDTYEDDDNVHLVMELCEGGELFDRIVAK--GHYTERAAATVVKTIVQVVQMCH 744
Query: 137 EQGVIHRDIKGANILTTKE---GLVKLADFGVATKLTEADINTHSVVGTPYWMAPEVIEM 193
E GV+HRD+K N L + +K DFG++ D + +VG+PY+MAPEV++
Sbjct: 745 EHGVMHRDLKPENFLFANKKETSPLKAIDFGLSITFKPGD-KFNEIVGSPYYMAPEVLKR 921
Query: 194 SGVCAASDIWSVGCTVIELLTCVPPYY---DLQPMPALFRIVQDEQPPIPDSLSPNITDF 250
+ DIWS G + LL +PP++ + A+ R V D + +S N D
Sbjct: 922 N-YGPEIDIWSAGVILYILLCGIPPFWAETEQGIAQAIIRSVIDFKKEPWPKVSDNAKDL 1098
Query: 251 LHQCFKKDARQRPDAKTLLSHPWILNCRRVLQSSL 285
+ + D ++R A+ +L HPW+ N + SL
Sbjct: 1099IKKMLDPDPKRRLTAQEVLDHPWLQNAKTAPNVSL 1203
>TC88402 similar to GP|20804521|dbj|BAB92215. putative CRK1 protein {Oryza
sativa (japonica cultivar-group)}, partial (46%)
Length = 2184
Score = 120 bits (302), Expect = 3e-27
Identities = 72/214 (33%), Positives = 120/214 (55%), Gaps = 7/214 (3%)
Frame = +1
Query: 25 EIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNIVKYLG 84
++GKG Y VYK D+ N VA+K+V +N+ E + + I+IL+ L+H NI+K G
Sbjct: 991 KLGKGTYSTVYKARDVTNQKIVALKRVRFDNLDPESVKFMAREIHILRRLDHPNIIKLEG 1170
Query: 85 SL--KTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQGVIH 142
+ +T L+++ EY+E+ P+ F E + Y+ Q+L GL + H GV+H
Sbjct: 1171 LITSETSRSLYLVFEYMEHDLTGLASNPSI--KFSEPQLKCYMHQLLSGLDHCHSHGVLH 1344
Query: 143 RDIKGANILTTKEGLVKLADFGVATKL-TEADINTHSVVGTPYWMAPE-VIEMSGVCAAS 200
RDIKG+N+L G++K+ADFG+A +I S V T ++ PE ++ + A
Sbjct: 1345 RDIKGSNLLIDNNGVLKIADFGLANVFDAHLNIPLTSRVVTLWYRPPELLLGANHYGVAV 1524
Query: 201 DIWSVGCTVIELLT---CVPPYYDLQPMPALFRI 231
D+WS GC + EL T +P +++ + +F++
Sbjct: 1525 DLWSTGCILRELYTGRPILPGKTEVEQLHRIFKL 1626
>TC78370 similar to GP|5001828|gb|AAD37165.1| 3-phosphoinositide-dependent
protein kinase-1 {Arabidopsis thaliana}, partial (91%)
Length = 2137
Score = 120 bits (301), Expect = 4e-27
Identities = 93/344 (27%), Positives = 157/344 (45%), Gaps = 16/344 (4%)
Frame = +1
Query: 20 YMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINI-LQNLNHKN 78
+ LG G G+Y +V + + G A+K + + I +E+ +++ I L L+H
Sbjct: 475 FELGKIYGVGSYSKVVRAKKKDTGVVYAMKIMDKKFITKENKTAYVKLERIVLDQLDHPG 654
Query: 79 IVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQ 138
IV+ + + L++ LE E G L + I + + Y A+V++ L Y+H
Sbjct: 655 IVRLYFTFQDTFSLYMALESCEGGELFDQI--TRKSRLTQDEAQFYAAEVVDALEYIHSL 828
Query: 139 GVIHRDIKGANILTTKEGLVKLADFGVATKLTEADI----------NTHSVVGTPYWMAP 188
GVIHRDIK N+L T EG +K+ADFG + ++ I + VGT ++ P
Sbjct: 829 GVIHRDIKPENLLLTTEGHIKIADFGSVKPMQDSQITVLPNAASDDKACTFVGTAAYVPP 1008
Query: 189 EVIEMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPIPDSLSPNIT 248
EV+ S +D+W++GCT+ ++L+ P+ D RI+ E PD S
Sbjct: 1009EVLNSSPATFGNDLWALGCTLYQMLSGTSPFKDASEWLIFQRIIARE-IRFPDYFSDEAR 1185
Query: 249 DFLHQCFKKDARQRPDA-----KTLLSHPWILNCRRVLQSSLRHSGTLRTIKDDGSAVAE 303
D + + D +RP A L SHP+ + V ++LR + + G+ +
Sbjct: 1186DLIDRLLDLDPSRRPGAGPDGYAILKSHPFF---KGVDWNNLRAQTPPKLALEPGAGESS 1356
Query: 304 VSGGDHKSTGEGSSVEKEDSAKEFSTGEANSRKSHEDNASDSNF 347
S H G +S + D A S G + + ++ DS +
Sbjct: 1357WSPA-HTGDGSAASARQPDGATSSSEGAGSITRLASIDSFDSKW 1485
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.132 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,544,686
Number of Sequences: 36976
Number of extensions: 534490
Number of successful extensions: 4078
Number of sequences better than 10.0: 611
Number of HSP's better than 10.0 without gapping: 3442
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3528
length of query: 1406
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1298
effective length of database: 5,021,319
effective search space: 6517672062
effective search space used: 6517672062
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0135.8


