

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0135.3
(361 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
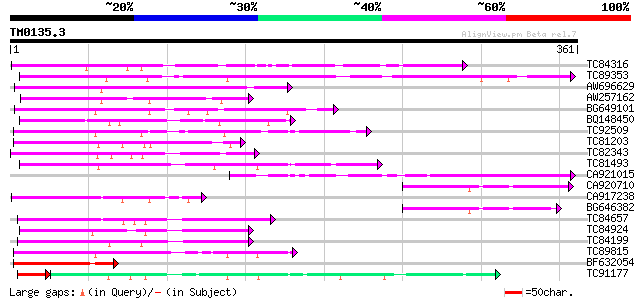
Score E
Sequences producing significant alignments: (bits) Value
TC84316 weakly similar to GP|11994612|dbj|BAB02749. gb|AAC24186.... 145 2e-35
TC89353 weakly similar to GP|9279704|dbj|BAB01261.1 gb|AAF25964.... 119 2e-27
AW696629 113 1e-25
AW257162 100 1e-21
BG649101 homologue to GP|6648199|gb|A unknown protein {Arabidops... 97 1e-20
BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.1... 89 3e-18
TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein... 88 6e-18
TC81203 weakly similar to GP|10177464|dbj|BAB10855. gb|AAD21700.... 82 4e-16
TC82343 similar to GP|10177628|dbj|BAB10775. gb|AAF30317.1~gene_... 79 4e-15
TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [i... 77 1e-14
CA921015 homologue to SP|Q8Y7G1|DPO3 DNA polymerase III polC-typ... 75 4e-14
CA920710 similar to PIR|S64314|S643 probable membrane protein YG... 75 4e-14
CA917238 weakly similar to PIR|A84538|A845 hypothetical protein ... 73 1e-13
BG646382 73 1e-13
TC84657 weakly similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.... 72 3e-13
TC84924 weakly similar to PIR|F96545|F96545 hypothetical protein... 72 3e-13
TC84199 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 70 2e-12
TC89815 69 3e-12
BF632054 similar to GP|10177628|dbj gb|AAF30317.1~gene_id:K6M13.... 68 6e-12
TC91177 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 57 6e-11
>TC84316 weakly similar to GP|11994612|dbj|BAB02749.
gb|AAC24186.1~gene_id:MGL6.4~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (10%)
Length = 892
Score = 145 bits (367), Expect = 2e-35
Identities = 110/305 (36%), Positives = 160/305 (52%), Gaps = 15/305 (4%)
Frame = +3
Query: 2 EPPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLH-----LHRSSSTTR 56
+P FLPEEL + IL LPV+SL+RF+CV KSWK++ SD+ F H ++
Sbjct: 27 KPLPFLPEELIVIILLRLPVRSLLRFKCVCKSWKTLFSDTHFANNHFLISTVYPQLVACE 206
Query: 57 NTDFAYLQSLITSPRKS--RSASTIAIP------DDYDFCGTCNGLVCLHSSKYDRKDKY 108
+ + T P +S ++ST IP Y G+CNG +CL+ D Y
Sbjct: 207 SVSAYRTWEIKTYPIESLLENSSTTVIPVSNTGHQRYTILGSCNGFLCLY-------DNY 365
Query: 109 TSHVRLWNPAMRSMSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVALSWDTLETMV 168
VRLWNP++ S+ SP + I Y+GF YD YK++AV A S T ETM
Sbjct: 366 QRCVRLWNPSINLKSKSSPTIDRFI----YYGFGYDQVNHKYKLLAVK-AFSRIT-ETM- 524
Query: 169 NVYNIGDDTCWRTIPVSHLPSM-YLKDDDAVYVNNTLNRLAILPNVEDRD-RCTILSFDF 226
+Y G+++C + + V P + +V+ TLN + V++RD R TILSFD
Sbjct: 525 -IYTFGENSC-KNVEVKDFPRYPPNRKHLGKFVSGTLNWI-----VDERDGRATILSFDI 683
Query: 227 GKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMHFVVWQMKEFGLYKSWT 286
KE+ Q+ L PQ Y Y P L VL +C+C+C + + +W MK++G+ +SWT
Sbjct: 684 EKETYRQVLL---PQHGYAVYS--PGLYVLSNCICVCTSFLDTRWQLWMMKKYGVAESWT 848
Query: 287 LLFNI 291
L +I
Sbjct: 849 KLMSI 863
>TC89353 weakly similar to GP|9279704|dbj|BAB01261.1
gb|AAF25964.1~gene_id:MYA6.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1482
Score = 119 bits (297), Expect = 2e-27
Identities = 113/380 (29%), Positives = 169/380 (43%), Gaps = 26/380 (6%)
Frame = +3
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDF------ 60
+PEE+ +EIL LPV+SL++FRCV K WK++ISD QF K H+ S++ +
Sbjct: 105 MPEEIIVEILLRLPVRSLLQFRCVCKLWKTLISDPQFAKKHVSISTAYPQLVSVFVSIAK 284
Query: 61 AYLQSLITSPRKSRSASTIAIPDDYD--------FCGTCNGLVCLHSSKYDRKDKYTSHV 112
L S P ++ P D++ G+CNGL+CL D Y
Sbjct: 285 CNLVSYPLKPLLDNPSAHRVEPADFEMIHTTSMTIIGSCNGLLCL-------SDFY--QF 437
Query: 113 RLWNPAMRSMSQRSPPLYLPIVYDS----YFGFDYDSSTDTYKVVAVAV-ALSWDTLETM 167
LWNP+++ S+ SP + +DS Y GF YD D YKV+AV + D +T+
Sbjct: 438 TLWNPSIKLKSKPSPTIIAFDSFDSKRFLYRGFGYDQVNDRYKVLAVVQNCYNLDETKTL 617
Query: 168 VNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILSFDFG 227
+ + D T + P + +V+ LN + + I+ FD
Sbjct: 618 IYTFGGKDWTTIQKFPCDPSRCDLGRLGVGKFVSGNLNWIV--------SKKVIVFFDIE 773
Query: 228 KESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKN-MHFVVWQMKEFGLYKSWT 286
KE+ ++ LP D+ L V + + + N H+VVW MKE+G+ +SWT
Sbjct: 774 KETYGEMSLP------QDYGDKNTVLYVSSNRIYVSFDHSNKTHWVVWMMKEYGVVESWT 935
Query: 287 LLFNIAGYAYDIP---CRFFAMYMSENGDALLL---SKLAVSQPQAVLYTQKDNKLEVTD 340
L I P C A+++SE+G L+ SKL+V D L+
Sbjct: 936 KLMIIPQDKLTSPGPYCLSDALFISEHGVLLMRPQHSKLSVYN------LNNDGGLDYCT 1097
Query: 341 VANYIFNCYAMNYIESLVPP 360
+ F Y Y ESLV P
Sbjct: 1098TISGQFARYLHIYNESLVSP 1157
>AW696629
Length = 663
Score = 113 bits (283), Expect = 1e-25
Identities = 69/192 (35%), Positives = 92/192 (46%), Gaps = 15/192 (7%)
Frame = +2
Query: 4 PQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRN------ 57
P LP +L ++ILSWLPVK L+RF VSK WKS+I D F KLHL +S T
Sbjct: 56 PPILPSDLIMQILSWLPVKLLIRFTSVSKHWKSLILDPNFAKLHLQKSPKNTHMILTALD 235
Query: 58 --------TDFAYLQSLITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYT 109
T + L+ S Y G+ NGLVCL K KY
Sbjct: 236 DEDDTWVVTPYPVRSLLLEQSSFSDEECCCFDYHSYFIVGSTNGLVCLAVEKSLENRKYE 415
Query: 110 SHVRLWNPAMRSMSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVALSWD-TLETMV 168
++ WNP++R S+++P L + + + GF YD DTYK AVA+ WD T M
Sbjct: 416 LFIKFWNPSLRLRSKKAPSLNIGLYGTARLGFGYDDLNDTYK----AVAVFWDHTTHKME 583
Query: 169 NVYNIGDDTCWR 180
+ D+CW+
Sbjct: 584 GRVHCMGDSCWK 619
>AW257162
Length = 728
Score = 100 bits (248), Expect = 1e-21
Identities = 61/167 (36%), Positives = 95/167 (56%), Gaps = 19/167 (11%)
Frame = +1
Query: 8 PEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRN-------TDF 60
P+++ EILSWL VK+L++ +CVSKSW ++ISDS F+K+HL+RS+ +++ D+
Sbjct: 130 PDDIIAEILSWLTVKTLMKMKCVSKSWNTLISDSNFVKMHLNRSARHSQSYLVSEHRGDY 309
Query: 61 AYLQSLITSPRKSRSASTIAIPDDYDF----------CGTCNGLVCLHSSKYDRKDKYTS 110
++ + RS I +P D + G+CNGLVCL D ++
Sbjct: 310 NFVPFSVRGLMNGRS---ITLPKDPYYQLIEKDCPGVVGSCNGLVCLSGCVADVEEFEEM 480
Query: 111 HVRLWNPAMRSMSQRSPPLYLPI--VYDSYFGFDYDSSTDTYKVVAV 155
+R+WNPA R++S + LY + F F YD++T TYKVVA+
Sbjct: 481 WLRIWNPATRTISDK---LYFSANRLQCWEFMFGYDNTTQTYKVVAL 612
>BG649101 homologue to GP|6648199|gb|A unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 799
Score = 96.7 bits (239), Expect = 1e-20
Identities = 71/234 (30%), Positives = 118/234 (50%), Gaps = 28/234 (11%)
Frame = +1
Query: 4 PQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSS---------- 53
P +P +L IL++LPVK++ + + VSKSW ++I+ FIK+HL++SS
Sbjct: 76 PVVIPSDLIAIILTFLPVKTITQLKLVSKSWNTLITSPSFIKIHLNQSSQNPNFILTPSR 255
Query: 54 ---TTRNTDFAYLQSLITSPRKSRSASTIAIPDDYDF--CGTCNGLVCLHSSKYDRKDKY 108
+ N + L+T S + +D+ F G+CNGL+CL K ++
Sbjct: 256 KQYSINNVLSVPIPRLLTGNTVSGDTYHNILNNDHHFRVVGSCNGLLCLLF-----KSEF 420
Query: 109 TSHV----RLWNPAMRSMSQ------RSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVA 158
+H+ R+WNPA R++S+ + PL+ + S F F D TYK+VA+
Sbjct: 421 ITHLKFRFRIWNPATRTISEELGFFRKYKPLFGGV---SRFTFGCDYLRGTYKLVALHTV 591
Query: 159 LSWDTLETMVNVYNIGD---DTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAI 209
D + + V V+N+G+ D CWR IP + + D V+++ T N L++
Sbjct: 592 EDGDVMRSNVRVFNLGNDDSDKCWRNIPNPFVCA------DGVHLSGTGNWLSL 735
>BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.18~similar to
unknown protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 88.6 bits (218), Expect = 3e-18
Identities = 69/198 (34%), Positives = 103/198 (51%), Gaps = 22/198 (11%)
Frame = +3
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAY---- 62
LP E+++EILS LPVK L++F+CV K WKS IS F+K HL R S+T +
Sbjct: 3 LPFEIQVEILSRLPVKYLMQFQCVCKLWKSQISKPDFVKKHL-RVSNTRHLFLLTFSKLS 179
Query: 63 ---------LQSLIT--SPRKSRSASTIAIPDDYD-FCGTCNGLVCLHSSKYDRKDKYTS 110
L S+ T +P ++ + D+ D G+C+G++C+ + S
Sbjct: 180 PELVIKSYPLSSVFTEMTPTFTQLEYPLNNRDESDSMVGSCHGILCIQCN--------LS 335
Query: 111 HVRLWNPAMRSMSQRSPPLYLP--IVYDSYFGFDYDSSTDTYKVVAVAVALSWDT----L 164
LWNP++R + + P P + + F YD S+DTYKVVAV + D L
Sbjct: 336 FPVLWNPSIRKFT-KLPSFEFPQNKFINPTYAFGYDHSSDTYKVVAVFCTSNIDNGVYQL 512
Query: 165 ETMVNVYNIGDDTCWRTI 182
+T+VNV+ +G + CWR I
Sbjct: 513 KTLVNVHTMGTN-CWRRI 563
>TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein F26G5.80 -
Arabidopsis thaliana, partial (8%)
Length = 858
Score = 87.8 bits (216), Expect = 6e-18
Identities = 79/250 (31%), Positives = 115/250 (45%), Gaps = 22/250 (8%)
Frame = +3
Query: 3 PPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSS----TTRNT 58
P LP EL EIL LPVK L + RC+ KS+ S+ISD +F K HLH S++ R+
Sbjct: 162 PLPTLPFELVAEILCRLPVKLLXQLRCLCKSFNSLISDPKFAKKHLHSSTTPHHLILRSN 341
Query: 59 DFAYLQSLITSPRKS-RSASTIAIP---------------DDYDFCGTCNGLVCLHSSKY 102
+ + +LI SP +S S ST+ +P Y++C +C+G++CL +
Sbjct: 342 NGSGRFALIVSPIQSVLSTSTVPVPQTQLTYPTCLTEEFASPYEWC-SCDGIICLTTD-- 512
Query: 103 DRKDKYTSHVRLWNPAMRSMSQRSPPLYLPIVY--DSYFGFDYDSSTDTYKVVAVAVALS 160
Y+S V LWNP + P Y+ + F F YD D YKV A+ +
Sbjct: 513 -----YSSAV-LWNPFINKFKTLPPLKYISLKRSPSCLFTFGYDPFADNYKVFAITFCVK 674
Query: 161 WDTLETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCT 220
T V V+ +G + WR I PS D ++V ++ L R+
Sbjct: 675 ----RTTVEVHTMGTSS-WRRI--EDFPSWSFIPDSGIFVAGYVHWLTYDGPGSQRE--- 824
Query: 221 ILSFDFGKES 230
I+S D ES
Sbjct: 825 IVSLDLEDES 854
>TC81203 weakly similar to GP|10177464|dbj|BAB10855.
gb|AAD21700.1~gene_id:MQB2.18~similar to unknown protein
{Arabidopsis thaliana}, partial (9%)
Length = 715
Score = 81.6 bits (200), Expect = 4e-16
Identities = 63/182 (34%), Positives = 86/182 (46%), Gaps = 34/182 (18%)
Frame = +1
Query: 3 PPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIIS-DSQFIKLHLHRSSSTT------ 55
P L +EL ++ILS LPVK+L++F+CV KSWK++IS D F KLHL RS T
Sbjct: 187 PTSLLLDELIVDILSRLPVKTLMQFKCVCKSWKTLISHDPSFAKLHLQRSPRNTHLTLVS 366
Query: 56 -RNTDFAYLQSLITSP--------------RKSRSASTIAIPDD-------YDFC---GT 90
R++D S++ P +AIPDD D C G+
Sbjct: 367 DRSSDDESNFSVVPFPVSHLIEAPLTITPFEPYHLLRNVAIPDDPYYVLGNMDCCIIIGS 546
Query: 91 CNGLVCLHSSKYDRKDKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDSYF--GFDYDSSTD 148
CNGL CL +++ R WNPA ++S+ L + +F F YD S D
Sbjct: 547 CNGLYCLRCYSLIYEEEEHDWFRFWNPATNTLSEELGCL------NEFFRLTFGYDISND 708
Query: 149 TY 150
TY
Sbjct: 709 TY 714
>TC82343 similar to GP|10177628|dbj|BAB10775.
gb|AAF30317.1~gene_id:K6M13.17~similar to unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 78.6 bits (192), Expect = 4e-15
Identities = 55/175 (31%), Positives = 86/175 (48%), Gaps = 16/175 (9%)
Frame = +2
Query: 1 MEPPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSST------ 54
++ P +LP+EL +IL LPVKSL+RF+ V KSW S+ISD+ F H +++T
Sbjct: 47 VDAPPYLPDELITKILVRLPVKSLIRFKSVCKSWFSLISDNHFANSHFQVTAATHTLRIL 226
Query: 55 --TRNTDFAYL--QSLITSPRKSRSA--STIAIPD---DYDFCGTCNGLVCLHSSKYDRK 105
T +F Y+ SL T +P+ D + +C G + +H+
Sbjct: 227 FLTATPEFRYIAVDSLFTDDYNEPVPLNPNFPLPEFEFDLEIKASCRGFIYVHT------ 388
Query: 106 DKYTSHVRLWNPAMRSMSQ-RSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVAL 159
S +WNP+ + Q PP ++ ++GF YD STD Y VV+V + +
Sbjct: 389 ---CSEAYIWNPSTGFLKQIPFPPNVSNLI---FYGFGYDESTDDYLVVSVYLVI 535
>TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 728
Score = 77.0 bits (188), Expect = 1e-14
Identities = 71/250 (28%), Positives = 117/250 (46%), Gaps = 19/250 (7%)
Frame = +1
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTT---RNTDFAYL 63
LP E+ EILS +P K L+R R K W+++I + FI LHL +S + R Y
Sbjct: 16 LPTEVTTEILSRVPAKPLLRLRSTCKWWRNLIDSTDFIFLHLSKSRDSVIILRQHSRLYE 195
Query: 64 QSLITSPR-KSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSHVRLWNPAMRS- 121
L + R K + + G+CNGL+C+ + D + WNP +R
Sbjct: 196 LDLNSMDRVKELDHPLMCYSNRIKVLGSCNGLLCICNIADD--------IAFWNPTIRKH 351
Query: 122 MSQRSPPL----------YLPIVYDSYFGFDYDSSTDTYKVVAVA--VALSWDTLETMVN 169
S PL ++ +GF YDS+TD YK+V+++ V L + ++ V
Sbjct: 352 RIIPSEPLIRKETNENNTITTLLAAHVYGFGYDSATDDYKLVSISNFVDLHNRSYDSHVT 531
Query: 170 VYNIGDDTCWRTIPVSHLP-SMYLKDDDAVYVNNTLNRLAILPN-VEDRDRCTILSFDFG 227
+Y +G D W +P+ +P ++ V+V+ L+ ++P +E R I++FD
Sbjct: 532 IYTMGSDV-W--MPLPGVPYALCCARPMGVFVSGALH--WVVPRALEPDSRDLIVAFDLR 696
Query: 228 KESCAQLPLP 237
E ++ LP
Sbjct: 697 FEVFREVALP 726
>CA921015 homologue to SP|Q8Y7G1|DPO3 DNA polymerase III polC-type (EC
2.7.7.7) (PolIII). {Listeria monocytogenes}, partial
(0%)
Length = 824
Score = 75.1 bits (183), Expect = 4e-14
Identities = 68/221 (30%), Positives = 102/221 (45%), Gaps = 1/221 (0%)
Frame = -1
Query: 141 FDYDSSTDTYKVVAVAVALSWDTLETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYV 200
F YD D +V+ V + V+VY +G D W+ I +P Y ++ V+V
Sbjct: 821 FGYDHFIDNIRVMFVLP-------KNEVSVYTLGTDY-WKRI--EDIP--YNIFEEGVFV 678
Query: 201 NNTLNRLAILPNVEDRDRCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCL 260
+ T+N LA D ILS D KES Q+ LP + D + LGVLR+CL
Sbjct: 677 SGTVNWLA-------SDDSFILSLDVEKESYQQVLLP---DTENDLW----ILGVLRNCL 540
Query: 261 CICQKDKNMHFVVWQMKEFGLYKSWTLLFNIAGYAYDIPCRFFAMYMSENGDALL-LSKL 319
CI N+ VW M E+G +SWT L+++ + +Y SE+ LL +++
Sbjct: 539 CILATS-NLFLDVWIMNEYGNQESWTKLYSVPNMQDHGLEAYTVLYSSEDDQLLLEFNEM 363
Query: 320 AVSQPQAVLYTQKDNKLEVTDVANYIFNCYAMNYIESLVPP 360
+ + V+Y K L + + N YIESL+ P
Sbjct: 362 RSDKVKLVVYDSKTGTLNIPEFQNNYDQICQNFYIESLISP 240
>CA920710 similar to PIR|S64314|S643 probable membrane protein YGR023w -
yeast (Saccharomyces cerevisiae), partial (7%)
Length = 774
Score = 75.1 bits (183), Expect = 4e-14
Identities = 43/122 (35%), Positives = 64/122 (52%), Gaps = 13/122 (10%)
Frame = -2
Query: 251 PTLGVLRDCLCICQKDKNMHFVVWQMKEFGLYKSWTLLFNI-------------AGYAYD 297
P L VL DCLC FV+WQMKEFG+ +SWT LF I Y +
Sbjct: 761 PYLRVLLDCLCFLHDLGKTEFVIWQMKEFGVRESWTRLFKIPYVDLQMLNLPIDVQYLNE 582
Query: 298 IPCRFFAMYMSENGDALLLSKLAVSQPQAVLYTQKDNKLEVTDVANYIFNCYAMNYIESL 357
P +Y+S+NGD ++L+ ++Y ++D +++ ++N I AM+Y+ESL
Sbjct: 581 YP--MLPLYISKNGDKVILTN--EKDDATIIYNKRDKRVDRARISNEIHWFSAMDYVESL 414
Query: 358 VP 359
VP
Sbjct: 413 VP 408
>CA917238 weakly similar to PIR|A84538|A845 hypothetical protein At2g16220
[imported] - Arabidopsis thaliana, partial (6%)
Length = 603
Score = 73.2 bits (178), Expect = 1e-13
Identities = 53/144 (36%), Positives = 72/144 (49%), Gaps = 20/144 (13%)
Frame = +2
Query: 2 EPPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFA 61
EPP LP+EL E+LS VKSL+R RCVS+ W S+ISD +F+KLH+ RS+ T +
Sbjct: 131 EPPSVLPDELITEVLSRGDVKSLMRMRCVSEYWNSMISDPRFVKLHMKRSARNAHLT-LS 307
Query: 62 YLQSLITSP--------RKSRSASTIAIPDDYDF----------CGTCNGLVCLHSSKYD 103
+S I R I +P D + G+CNG +CL +
Sbjct: 308 LCKSGIDGDNNVVPYPVRGLIENGLITLPSDPYYRLKDKECQYVIGSCNGWLCLLG--FS 481
Query: 104 RKDKYTSHV--RLWNPAMRSMSQR 125
Y H+ R WNPAM M+Q+
Sbjct: 482 SIGAY-RHIWFRFWNPAMGKMTQK 550
>BG646382
Length = 603
Score = 73.2 bits (178), Expect = 1e-13
Identities = 46/120 (38%), Positives = 62/120 (51%), Gaps = 19/120 (15%)
Frame = +3
Query: 251 PTLGVLRDCLCICQKDKNMHFVVWQMKEFGLYKSWTLLFNI------------------A 292
P+L VL DCLC K FV+WQMKEFG +SWT LF I
Sbjct: 231 PSLRVLMDCLCFSHDFKRTEFVIWQMKEFGSQESWTQLFRIKYINLQIHNLPINDNLDLL 410
Query: 293 GY-AYDIPCRFFAMYMSENGDALLLSKLAVSQPQAVLYTQKDNKLEVTDVANYIFNCYAM 351
GY +IP +Y+SENGD L+LS + ++Y Q+D ++E T ++N I C A+
Sbjct: 411 GYMECNIP--LLPLYLSENGDTLILSN--DEEEGVIIYNQRDKRVEKTRISNEI--CLAL 572
>TC84657 weakly similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 884
Score = 72.0 bits (175), Expect = 3e-13
Identities = 58/182 (31%), Positives = 85/182 (45%), Gaps = 18/182 (9%)
Frame = +1
Query: 6 FLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQS 65
+LP EL ++I+ LPVKSL+RF+CV KS ++ISD F K H S++T N ++ +
Sbjct: 325 YLPHELIIQIMLRLPVKSLIRFKCVCKSLLALISDHNFAKSHFELSTATHTNR-IVFMST 501
Query: 66 LITSPR--------KSRSAST-----IAIPDDY---DFCGTCNGLVCLHSSKYDRKDKYT 109
L R SAST P+ Y + +C G + L S
Sbjct: 502 LALETRSIDFEASLNDDSASTSLNLNFMPPESYSSLEIKSSCRGFIVLTCS--------- 654
Query: 110 SHVRLWNPAMRSMSQ-RSPPLYLPIVYD-SYFGFDYDSSTDTYKVVAVAVALSWDTLETM 167
S++ LWNP+ Q P L Y +GF YD D Y VV+V+ S D ++
Sbjct: 655 SNIYLWNPSTGHHKQIPFPASNLDAKYSCCLYGFGYDHLRDDYLVVSVSYNTSIDPVDDN 834
Query: 168 VN 169
++
Sbjct: 835 IS 840
>TC84924 weakly similar to PIR|F96545|F96545 hypothetical protein F8A12.9
[imported] - Arabidopsis thaliana, partial (9%)
Length = 575
Score = 72.0 bits (175), Expect = 3e-13
Identities = 52/167 (31%), Positives = 76/167 (45%), Gaps = 18/167 (10%)
Frame = +1
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRS--------------S 52
LP+EL ++ L LPVKSL+ F+C+ K W SIISD F H + S
Sbjct: 61 LPQELIIQFLLRLPVKSLLVFKCICKLWFSIISDPHFANSHFQLNHAKHTRRFLCISALS 240
Query: 53 STTRNTDF-AYLQSLITSPRKSRSASTIAIPDDY---DFCGTCNGLVCLHSSKYDRKDKY 108
R+ DF A+L SP + S +PD Y + G+C G + ++
Sbjct: 241 PEIRSIDFDAFLNDAPASPNFNCS-----LPDSYFPFEIKGSCRGFIFMYRH-------- 381
Query: 109 TSHVRLWNPAMRSMSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAV 155
++ +WNP+ S Q + Y + +GF YD S D Y VV +
Sbjct: 382 -PNIYIWNPSTGSKRQILMSAFNTKAYINLYGFGYDQSRDDYVVVFI 519
>TC84199 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 673
Score = 69.7 bits (169), Expect = 2e-12
Identities = 52/166 (31%), Positives = 73/166 (43%), Gaps = 16/166 (9%)
Frame = +1
Query: 6 FLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAY--- 62
+LP EL ++IL WLPVKSL+RF+CV KSW S+ISD+ F H ++ +R F
Sbjct: 106 YLPLELIIQILLWLPVKSLLRFKCVCKSWFSLISDTHFANSHFQITAKHSRRVLFMLNHV 285
Query: 63 ---LQSLITSPRKSRSASTIAIP---------DDYDF-CGTCNGLVCLHSSKYDRKDKYT 109
L + + S I P D D +C G + LH+
Sbjct: 286 PTTLSLDFEALHCDNAVSEIPNPIPNFVEPPCDSLDTNSSSCRGFIFLHNDP-------- 441
Query: 110 SHVRLWNPAMRSMSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAV 155
+ +WNP+ R Q + +GF YD D Y VV+V
Sbjct: 442 -DLFIWNPSTRVYKQIPLSPNDSNSFHCLYGFGYDQLRDDYLVVSV 576
>TC89815
Length = 807
Score = 68.9 bits (167), Expect = 3e-12
Identities = 58/197 (29%), Positives = 90/197 (45%), Gaps = 16/197 (8%)
Frame = +1
Query: 3 PPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSS----TTRNT 58
P +P E+ ++ILS LP L+RFR SKS KSII F LHL S++ N
Sbjct: 34 PMANIPPEIFIDILSLLPPHPLLRFRSTSKSLKSIIDSHTFTNLHLKNSNNFYLIIRHNA 213
Query: 59 DFAYLQSLITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSHVRLWNPA 118
+ L +P + ++ + G+CNGL+C+ + D + WNP
Sbjct: 214 NLYQLDFPNLTPPIPLNHPLMSYSNRITLFGSCNGLICISNIADD--------IAFWNPN 369
Query: 119 MRSMSQRSPPLYLPIVYDS----------YFGFDYDSSTDTYKVVAVA--VALSWDTLET 166
+R R P YLP S GF YDS YK+V ++ V L + ++
Sbjct: 370 IR--KHRIIP-YLPTTPRSESDTTLFAARVHGFGYDSFAGDYKLVRISYFVDLQNRSFDS 540
Query: 167 MVNVYNIGDDTCWRTIP 183
V V+++ ++ W+ +P
Sbjct: 541 QVRVFSLKMNS-WKELP 588
>BF632054 similar to GP|10177628|dbj gb|AAF30317.1~gene_id:K6M13.17~similar
to unknown protein {Arabidopsis thaliana}, partial (8%)
Length = 462
Score = 67.8 bits (164), Expect = 6e-12
Identities = 39/67 (58%), Positives = 44/67 (65%)
Frame = +3
Query: 3 PPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAY 62
P FLPEEL LEIL LP+KSL+RFRCV KSW IIS+ FIK LH S T+NT F
Sbjct: 132 PLPFLPEELILEILIKLPIKSLLRFRCVCKSWLHIISNPYFIKKQLHFS---TQNTHFTT 302
Query: 63 LQSLITS 69
+I S
Sbjct: 303 NHRIILS 323
>TC91177 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 1298
Score = 56.6 bits (135), Expect(2) = 6e-11
Identities = 75/329 (22%), Positives = 133/329 (39%), Gaps = 43/329 (13%)
Frame = +2
Query: 27 FRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFA---YLQSLITSPRKSRS-------- 75
F+CV K W S+IS F H +++T N YLQSL S +
Sbjct: 74 FKCVCKLWLSLISQPHFANSHFQLTTATHTNRIMLITPYLQSLSIDLELSLNDDSAVYTT 253
Query: 76 -ASTIAIPDDY--------------------DFCGTCNGLVCLHSSKYDRKDKYTSHVRL 114
S + +DY DF G+C G + L+ S + +
Sbjct: 254 DISFLIDDEDYYSSSSSDMDDLSPPKSFFILDFKGSCRGFILLNCY---------SSLCI 406
Query: 115 WNPAMRSMSQRSPPLYLPIVYDS--YFGFDYDSSTDTYKVVAVAV--ALSWDTLETMVNV 170
WNP+ +R P + D+ ++GF YD STD Y V++++ + S D + + + +
Sbjct: 407 WNPST-GFHKRIPFTTIDSNPDANYFYGFGYDESTDDYLVISMSYEPSPSSDGMLSHLGI 583
Query: 171 YNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAI--LPNVEDRDRCTILSFDFGK 228
+++ + W + +L + + + + +L+ AI L D I++F +
Sbjct: 584 FSLRANV-WTRVEGGNL--LLYSQNSLLNLVESLSNGAIHWLAFRNDISMPVIVAFHLME 754
Query: 229 ESCAQLPLP----YCPQSRYDFYRTWPTLGVLRDCLCICQ-KDKNMHFVVWQMKEFGLYK 283
+L LP P YD L V R CL + + F +W M+++ +
Sbjct: 755 RKLLELRLPNEIINGPSRAYD-------LWVYRGCLALWHILPDRVTFQIWVMEKYNVQS 913
Query: 284 SWTLLFNIAGYAYDIPCRFFAMYMSENGD 312
SWT + + + F+ Y +++GD
Sbjct: 914 SWTKTL-VLSFDGNPAHSFWPKYYTKSGD 997
Score = 27.7 bits (60), Expect(2) = 6e-11
Identities = 13/21 (61%), Positives = 17/21 (80%)
Frame = +3
Query: 6 FLPEELRLEILSWLPVKSLVR 26
+LP EL ++IL LPVKSL+R
Sbjct: 12 YLPFELIIQILLRLPVKSLIR 74
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.137 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,260,300
Number of Sequences: 36976
Number of extensions: 234492
Number of successful extensions: 1372
Number of sequences better than 10.0: 139
Number of HSP's better than 10.0 without gapping: 1306
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1322
length of query: 361
length of database: 9,014,727
effective HSP length: 97
effective length of query: 264
effective length of database: 5,428,055
effective search space: 1433006520
effective search space used: 1433006520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0135.3


