

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0135.13
(479 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
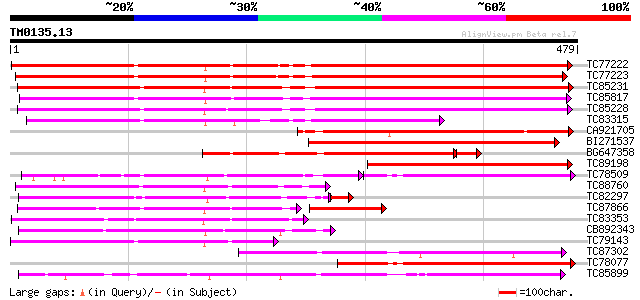
Score E
Sequences producing significant alignments: (bits) Value
TC77222 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 424 e-119
TC77223 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 398 e-111
TC85231 similar to GP|13508844|emb|CAC35167. arbutin synthase {R... 374 e-104
TC85817 weakly similar to PIR|B85014|B85014 probable flavonol gl... 365 e-101
TC85228 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 355 2e-98
TC83315 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 249 2e-66
CA921705 weakly similar to GP|13508844|em arbutin synthase {Rauv... 234 7e-62
BI271537 similar to GP|19911209|db glucosyltransferase-13 {Vigna... 233 1e-61
BG647358 similar to GP|19911209|db glucosyltransferase-13 {Vigna... 195 3e-54
TC89198 similar to GP|19911209|dbj|BAB86931. glucosyltransferase... 206 2e-53
TC78509 similar to GP|19911193|dbj|BAB86923. glucosyltransferase... 136 2e-53
TC88760 similar to GP|19911209|dbj|BAB86931. glucosyltransferase... 185 4e-47
TC82297 similar to GP|19911209|dbj|BAB86931. glucosyltransferase... 178 6e-47
TC87866 similar to GP|19911195|dbj|BAB86924. glucosyltransferase... 121 2e-45
TC83353 similar to GP|19911195|dbj|BAB86924. glucosyltransferase... 179 3e-45
CB892343 similar to GP|19911195|dbj glucosyltransferase-6 {Vigna... 176 2e-44
TC79143 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 166 2e-41
TC87302 similar to GP|19911199|dbj|BAB86926. glucosyltransferase... 164 5e-41
TC78077 similar to GP|4115536|dbj|BAA36411.1 UDP-glycose:flavono... 155 4e-38
TC85899 weakly similar to GP|7385017|gb|AAF61647.1| UDP-glucose:... 150 1e-36
>TC77222 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (76%)
Length = 1643
Score = 424 bits (1091), Expect = e-119
Identities = 221/480 (46%), Positives = 313/480 (65%), Gaps = 6/480 (1%)
Frame = +3
Query: 2 EVTTTTHIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLP 61
++ T HIA++ P +SH I+EF KRL H + H+TCI P++ S + K+ E +P
Sbjct: 18 KMAKTIHIAVIPSPGFSHLVPIVEFSKRLVTNHPNFHVTCIIPSLGSPPDSSKSYLEKIP 197
Query: 62 PNIDSMFLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDV 121
PNI+S+FLPP+N D+P+ + IL+ T+T SLPSI+ ALK+L S+ L +I+ D
Sbjct: 198 PNINSIFLPPINKQDLPQGVYPAILIQQTVTLSLPSIHQALKSL---SSKAPLVAIIADS 368
Query: 122 LITQVLPMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISCE------TVEIPGCMLIH 175
+ L A E N LSY YF ++A +LS L+ D+ +SCE +++ GC+ I+
Sbjct: 369 FAFEALDFAKEFNSLSYFYFPTSANVLSFILHLPKLDEEVSCEFKDLQEPIKLQGCVPIN 548
Query: 176 TTDLPDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTS 235
DLP P ++R SE Y++FL+ K FY DG++INSF++LE AL++ KG + S
Sbjct: 549 GIDLPTPTKDR--SSEAYRMFLQRAKSFYFVDGILINSFYELESSAVEALKQ-KGYGNIS 719
Query: 236 IPHVYPVGPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFN 295
+PVGP+ Q GS + G+ HE ++WL Q QNSVLYVSFGS GTL+ Q N
Sbjct: 720 Y---FPVGPITQIGS----SNNDVVGDEHECLKWLKNQPQNSVLYVSFGSGGTLSQRQIN 878
Query: 296 ELALGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWAD 355
E+A GLELSG +F+W +R P L +EDPL+FLP GF ERTK +GF++P WA
Sbjct: 879 EIAFGLELSGQRFIWGVRAPSDSVNAAYLESTNEDPLKFLPEGFQERTKEKGFILPSWAP 1058
Query: 356 QIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKP 415
Q+EIL H ++GGFL HCGWNS+LES G+PIVAWPL+A+Q M A + DGLKVA+R K
Sbjct: 1059QVEILKHSSVGGFLSHCGWNSVLESMQEGVPIVAWPLFAEQAMNAVMLCDGLKVAIRLKF 1238
Query: 416 NEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQLALKWK 475
+ IV + A VIK ++ GEEGK ++ RMK L+D A++A+K++GSS + ++QLA +W+
Sbjct: 1239EDDEIVEKDKTANVIKCLMEGEEGKTMRDRMKSLKDYAVNAVKDEGSSIQNLSQLASQWE 1418
>TC77223 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (50%)
Length = 1628
Score = 398 bits (1023), Expect = e-111
Identities = 213/473 (45%), Positives = 300/473 (63%), Gaps = 7/473 (1%)
Frame = +3
Query: 6 TTHIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNID 65
T HIA++ P +SH I+EF KRL H + H+TCI P++ S + K+ E +PPNI+
Sbjct: 21 TIHIAVIPSPGFSHLVPIVEFTKRLVTNHPNFHVTCIIPSLGSPPDSSKSYLETIPPNIN 200
Query: 66 SMFLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVLITQ 125
S+FLPP+N D+P+ H +L+ T+T SLPSI+ AL++L ++ L +I+ D +
Sbjct: 201 SIFLPPINKQDLPQGVHPGVLIQLTVTHSLPSIHQALESL---TSKTPLVAIIADTFAFE 371
Query: 126 VLPMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISCE------TVEIPGCMLIHTTDL 179
L A E N LSY+YF ++ +LSL L+ D+ SCE +++ GC+ I+ DL
Sbjct: 372 ALDFAKEFNSLSYLYFPCSSFVLSLLLHLPKLDEEFSCEYKDLQEPIKLQGCVPINGIDL 551
Query: 180 PDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTSIPHV 239
P ++R +E YK++++ K Y DG++INSF +LE +AL E KG
Sbjct: 552 PAATKDR--SNEGYKMYIQRAKSMYFVDGILINSFIELESSAIKAL-ELKGYGKIDF--- 713
Query: 240 YPVGPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELAL 299
+PVGP+ Q G S + G+ E ++WL Q QNSVLYVSFGS GTL+ Q NELA
Sbjct: 714 FPVGPITQTGL-----SNNDVGDELECLKWLKNQPQNSVLYVSFGSGGTLSQTQINELAF 878
Query: 300 GLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWADQIEI 359
GLELSG +F+WV+R P L +EDPL+FLP GFLERTK +G ++P WA Q++I
Sbjct: 879 GLELSGQRFIWVLRAPSDSVSAAYLEATNEDPLKFLPKGFLERTKEKGLILPSWAPQVQI 1058
Query: 360 LGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPNEKG 419
L ++GGFL HCGWNS+LES G+PIVAWPL+A+Q M A + S+ LKVA+R K +
Sbjct: 1059LKEKSVGGFLSHCGWNSVLESMQEGVPIVAWPLFAEQAMNAVMLSNDLKVAIRLKFEDDE 1238
Query: 420 IVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALK-EDGSSTRTITQLA 471
IV + +A VIK ++ GEEGK ++ RMK L+D A AL +DGSS +T++ LA
Sbjct: 1239IVEKDKIANVIKCLMEGEEGKAMRDRMKSLRDYATKALNVKDGSSIQTLSHLA 1397
>TC85231 similar to GP|13508844|emb|CAC35167. arbutin synthase {Rauvolfia
serpentina}, partial (34%)
Length = 1602
Score = 374 bits (959), Expect = e-104
Identities = 209/479 (43%), Positives = 293/479 (60%), Gaps = 9/479 (1%)
Frame = +3
Query: 7 THIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNIDS 66
T IA+V P SH ++EF K L H + HIT + PT+ ++++ LPPN++
Sbjct: 60 TCIAMVPSPGLSHLIPLVEFAKLLLQNHNEYHITFLIPTLGPLTPSMQSILNTLPPNMNF 239
Query: 67 MFLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVLITQV 126
+ LP VN++D+P + + S+P +Y +K+L + L ++V + T
Sbjct: 240 IVLPQVNIEDLPHNLDPATQMKLIVKHSIPFLYEEVKSLLSKTR---LVALVFSMFSTDA 410
Query: 127 LPMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISC-------ETVEIPGCML-IHTTD 178
+A N+LSY++FSS A+L SL L D+ S ETV IPG + +H +
Sbjct: 411 HDVAKHFNLLSYLFFSSGAVLFSLFLTIPNLDEAASTQFLGSSYETVNIPGFSIPLHIKE 590
Query: 179 LPDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTSIPH 238
LPDP R S+ YK L+ ++ L DGVI+N+F DLEPE R LQ+ + P
Sbjct: 591 LPDPFICE-RSSDAYKSILDVCQKLSLFDGVIMNTFTDLEPEVIRVLQDREK------PS 749
Query: 239 VYPVGPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELA 298
VYPVGP+++ S NE +RWL+ Q+ +SVL+VSFGS GTL+ DQ NE+A
Sbjct: 750 VYPVGPMIRNES-------NNEANMSMCLRWLENQQPSSVLFVSFGSGGTLSQDQLNEIA 908
Query: 299 LGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWADQIE 358
GLELSG KFLWV+R P K S + DPLE+LPNGFLERTK G VV WA Q+E
Sbjct: 909 FGLELSGHKFLWVVRAPSKNSSSAYFSGQNNDPLEYLPNGFLERTKENGLVVASWAPQVE 1088
Query: 359 ILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPK-PNE 417
ILGHG+IGGFL HCGW+S LES ++G+P++AWPL+A+Q+M A L +D LKVA+RPK +E
Sbjct: 1089ILGHGSIGGFLSHCGWSSTLESVVNGVPLIAWPLFAEQRMNAKLLTDVLKVAVRPKVDDE 1268
Query: 418 KGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQLALKWKR 476
GI+ + VA+ IK I+ G+E EI++++K L A L E GSS + ++ LALKW++
Sbjct: 1269TGIIKQEEVAKAIKRIMKGDESFEIRKKIKELSVGAATVLSEHGSSRKALSSLALKWQQ 1445
>TC85817 weakly similar to PIR|B85014|B85014 probable flavonol
glucosyltransferase [imported] - Arabidopsis thaliana,
partial (83%)
Length = 1736
Score = 365 bits (936), Expect = e-101
Identities = 206/475 (43%), Positives = 285/475 (59%), Gaps = 9/475 (1%)
Frame = +3
Query: 9 IAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNIDSMF 68
+ ++ P H ++EF KR+ L+Q++ IT PT K + ++LP I F
Sbjct: 162 VVMLPSPGMGHLIPMIEFAKRIIILNQNLQITFFIPTEGPPSKAQKTVLQSLPKFISHTF 341
Query: 69 LPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVLITQVLP 128
LPPV+ D+P I ++ T+ SLPS+ TL S + +T++VVD+ T
Sbjct: 342 LPPVSFSDLPPNSGIETIISLTVLRSLPSLRQNFNTL---SETHTITAVVVDLFGTDAFD 512
Query: 129 MANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISCE------TVEIPGCMLIHTTDLPDP 182
+A E NV YV++ STAM LSL LY D+ + CE V+IPGC+ IH L DP
Sbjct: 513 VAREFNVPKYVFYPSTAMALSLFLYLPRLDEEVHCEFRELTEPVKIPGCIPIHGKYLLDP 692
Query: 183 VQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRAL-QENKGTSSTSIPHVYP 241
+Q+R ++ Y+ KR+ ADG+I NSF +LEP + L +E G P YP
Sbjct: 693 LQDRK--NDAYQSVFRNAKRYREADGLIENSFLELEPGPIKELLKEEPGK-----PKFYP 851
Query: 242 VGPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGL 301
VGPLV+ G G E ++WLD Q SVL+VSFGS GTL+ Q ELALGL
Sbjct: 852 VGPLVKR-----EVEVGQIGPNSESLKWLDNQPHGSVLFVSFGSGGTLSSKQIVELALGL 1016
Query: 302 ELSGVKFLWVIRPPQ-KLEMIGDLSVGHE-DPLEFLPNGFLERTKGQGFVVPYWADQIEI 359
E+S +FLWV+R P K+ S + DP +FLPNGFLERTKG+G VV WA Q ++
Sbjct: 1017EMSEQRFLWVVRSPNDKVANASYFSAETDSDPFDFLPNGFLERTKGRGLVVSSWAPQPQV 1196
Query: 360 LGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPNEKG 419
L HG+ GGFL HCGWNS+LES ++G+P+V WPLYA+QKM A + ++ +KV LRP E G
Sbjct: 1197LAHGSTGGFLTHCGWNSVLESVVNGVPLVVWPLYAEQKMNAVMLTEDVKVGLRPNVGENG 1376
Query: 420 IVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQLALKW 474
+V R +A V+K ++ GEEGK+++ +MK L++ A L E+G+ST I+ LALKW
Sbjct: 1377LVERLEIASVVKCLMEGEEGKKLRYQMKDLKEAASKTLGENGTSTNHISNLALKW 1541
>TC85228 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (31%)
Length = 3139
Score = 355 bits (911), Expect = 2e-98
Identities = 201/478 (42%), Positives = 286/478 (59%), Gaps = 9/478 (1%)
Frame = -1
Query: 7 THIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNIDS 66
T IA+V P SH +EF K L H + HIT + PT+ ++++ LPPN++
Sbjct: 2776 TCIAMVPSPGLSHLIPQVEFAKLLLQHHNESHITFLIPTLGPLTPSMQSILNTLPPNMNF 2597
Query: 67 MFLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVLITQV 126
LP VN++D+P + + S+P ++ +K+L +N L ++V + T
Sbjct: 2596 TVLPQVNIEDLPHNLEPSTQMKLIVKHSIPFLHEEVKSLLSKTN---LVALVCSMFSTDA 2426
Query: 127 LPMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISC-------ETVEIPGCML-IHTTD 178
+A N+LSY++FSS A+L S L D S E V +PG + H +
Sbjct: 2425 HDVAKHFNLLSYLFFSSGAVLFSFFLTLPNLDDAASTQFLGSSYEMVNVPGFSIPFHVKE 2246
Query: 179 LPDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTSIPH 238
LPDP N R S+ YK L+ ++ L DGVIIN+F +LE E R LQ+ + P
Sbjct: 2245 LPDPF-NCERSSDTYKSILDVCQKSSLFDGVIINTFSNLELEAVRVLQDREK------PS 2087
Query: 239 VYPVGPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELA 298
V+PVGP+++ S NE +RWL+ Q +SV++VSFGS GTL+ DQ NELA
Sbjct: 2086 VFPVGPIIRNES-------NNEANMSVCLRWLENQPPSSVIFVSFGSGGTLSQDQLNELA 1928
Query: 299 LGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWADQIE 358
GLELSG KFLWV+R P K + + +PLE+LPNGF+ERTK +G VV WA Q+E
Sbjct: 1927 FGLELSGHKFLWVVRAPSKHSSSAYFNGQNNEPLEYLPNGFVERTKEKGLVVTSWAPQVE 1748
Query: 359 ILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPK-PNE 417
ILGHG+IGGFL HCGW+S LES ++G+P++AWPL+A+Q+M A L +D LKVA+RPK E
Sbjct: 1747 ILGHGSIGGFLSHCGWSSTLESVVNGVPLIAWPLFAEQRMNAKLLTDVLKVAVRPKVDGE 1568
Query: 418 KGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQLALKWK 475
GI+ R V++ +K I+ G+E EI++++K L A L E GSS + ++ LALKW+
Sbjct: 1567 TGIIKREEVSKALKRIMEGDESFEIRKKIKELSVSAATVLSEHGSSRKALSTLALKWQ 1394
>TC83315 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (42%)
Length = 1048
Score = 249 bits (636), Expect = 2e-66
Identities = 146/364 (40%), Positives = 204/364 (55%), Gaps = 11/364 (3%)
Frame = +2
Query: 15 PLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNIDSMFLPPVNL 74
P +SH I+EF KRL H + H+TCI P++ S + K+ E +PPNI+S+FLPP+N
Sbjct: 8 PGFSHLVPIVEFSKRLVTNHPNFHVTCIIPSLGSPPDSSKSYLETIPPNINSIFLPPINK 187
Query: 75 DDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVLITQVLPMANELN 134
D+P+ + IL+ T+T SLPSI+ ALK+L+ + L +I+ D+ + L A E N
Sbjct: 188 QDLPQGVYPAILIQQTVTLSLPSIHQALKSLNSKAP---LVAIIADIFAQETLDFAKEFN 358
Query: 135 VLSYVYFSSTAMLLSLCLYPSTFDKTISCE------TVEIPGCMLIHTTDLPDPVQNRFR 188
L Y+YF S+A +LSL L+ D+ +SCE +++ GC+ I+ DLP P ++R
Sbjct: 359 SLFYLYFPSSAFVLSLVLHIPNLDEEVSCEYKDLKEPIKLQGCLPINGIDLPTPTKDRSN 538
Query: 189 -----CSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTSIPHVYPVG 243
S K + G L N + + E KG +PVG
Sbjct: 539 ESLQNASSTCKKYAFG*WNLGLIAS*N*N-------QVLQKALEQKGYGKIGF---FPVG 688
Query: 244 PLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLEL 303
+ Q GS + G+ HE ++WL Q QNSVLYVSFGS GTL+ Q NELA GLEL
Sbjct: 689 TITQIGS----SNNDVVGDEHECLKWLKNQPQNSVLYVSFGSGGTLSQTQINELAFGLEL 856
Query: 304 SGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWADQIEILGHG 363
SG +F+WV+R P L +EDPL+FLP G LERTK +G + WA Q+EIL H
Sbjct: 857 SGQRFIWVVRAPSDSVSAAYLESTNEDPLKFLPIGXLERTKEKGXYLASWAPQVEILKHS 1036
Query: 364 AIGG 367
++GG
Sbjct: 1037SVGG 1048
>CA921705 weakly similar to GP|13508844|em arbutin synthase {Rauvolfia
serpentina}, partial (34%)
Length = 787
Score = 234 bits (596), Expect = 7e-62
Identities = 117/236 (49%), Positives = 168/236 (70%), Gaps = 3/236 (1%)
Frame = -2
Query: 244 PLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLEL 303
P++Q+ +C+ +++ + + WLD+Q+ NSV++VSFGS GT++ +Q NELALGLEL
Sbjct: 786 PIIQQ-NCDNTQNES------QCLSWLDEQKPNSVVFVSFGSGGTISQNQMNELALGLEL 628
Query: 304 SGVKFLWVIRPPQKLE--MIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWADQIEILG 361
S KFLWV+R P + + D+S +DPL FLP GFLERT QGF+V WA Q+EIL
Sbjct: 627 SSQKFLWVVREPNDIASAIYFDVSNSKKDPLSFLPKGFLERTNKQGFLVSNWAPQVEILS 448
Query: 362 HGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPK-PNEKGI 420
H AIGGF+ HCGW S LE ++G+PIVAWPL+A+Q+M A + +DG+K+A+RP N G+
Sbjct: 447 HKAIGGFVTHCGWFSTLECVVNGVPIVAWPLFAEQRMNATILADGIKIAIRPTIDNVSGV 268
Query: 421 VGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQLALKWKR 476
V + + V+K ++V +EG EI++RMK L+D A +A+K DGSS T++QL KW +
Sbjct: 267 VEKVEIVNVLKRLIV-DEGIEIRRRMKVLKDAAANAMKVDGSSIITMSQLVTKWTK 103
>BI271537 similar to GP|19911209|db glucosyltransferase-13 {Vigna angularis},
partial (34%)
Length = 652
Score = 233 bits (594), Expect = 1e-61
Identities = 119/214 (55%), Positives = 152/214 (70%), Gaps = 2/214 (0%)
Frame = +2
Query: 253 THESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLELSGVKFLWVI 312
T ES G+ E + WLDKQ+ SVLYVSFGS GTL+ +Q ELALGLELS FLWV+
Sbjct: 8 TIESSGDANHGLECLTWLDKQQPCSVLYVSFGSGGTLSQEQIVELALGLELSNKIFLWVL 187
Query: 313 RPPQKLEM-IGDLSVGHE-DPLEFLPNGFLERTKGQGFVVPYWADQIEILGHGAIGGFLC 370
R P G S ++ D +FLP+GFLERTK +GFV+ W QI+IL H ++GGFL
Sbjct: 188 RAPSSSSSSAGYFSAQNDADTWQFLPSGFLERTKEKGFVITSWVPQIQILSHNSVGGFLT 367
Query: 371 HCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPNEKGIVGRGVVAEVI 430
HCGWNS LES +HG+P++ WPL+A+QKM A L S+GLKV LR NE GIV R VA+VI
Sbjct: 368 HCGWNSTLESVVHGVPLITWPLFAEQKMNAVLLSEGLKVGLRASVNENGIVERVEVAKVI 547
Query: 431 KNILVGEEGKEIQQRMKRLQDVAIDALKEDGSST 464
K ++ GEEG++++ MK L++ A +A+KEDGSST
Sbjct: 548 KCLMEGEEGEKLRNNMKELKESASNAVKEDGSST 649
>BG647358 similar to GP|19911209|db glucosyltransferase-13 {Vigna angularis},
partial (33%)
Length = 738
Score = 195 bits (495), Expect(2) = 3e-54
Identities = 106/218 (48%), Positives = 140/218 (63%), Gaps = 2/218 (0%)
Frame = +3
Query: 164 ETVEIPGCMLIHTTDLPDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFR 223
E +++PGC+ +H DL VQ+R S+ YK FL+ K ADGV++NSF ++E
Sbjct: 57 EPIKVPGCVPLHGRDLLTIVQDR--SSQAYKYFLQHVKSLSFADGVLVNSFLEMEMGPIN 230
Query: 224 ALQENKGTSSTSIPHVYPVGPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSF 283
AL E + P VYPVGP++Q + ++ G E + WLDKQ+ SVLYVSF
Sbjct: 231 ALTEE----GSGNPSVYPVGPIIQTVTGSVDDANGLE-----CLSWLDKQQSCSVLYVSF 383
Query: 284 GSAGTLTHDQFNELALGLELSGVKFLWVIRPPQKLEM-IGDLSVGHE-DPLEFLPNGFLE 341
GS GTL+H+Q ELALGLELS KFLWV+R P LS ++ D L+FLP+GFLE
Sbjct: 384 GSGGTLSHEQIVELALGLELSNQKFLWVVRAPSSSSSNAAYLSAQNDVDALQFLPSGFLE 563
Query: 342 RTKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLE 379
RTK +GFV+ WA QI+IL H ++GGFL HCGW+S L+
Sbjct: 564 RTKEEGFVITSWAPQIQILSHSSVGGFLSHCGWSSHLK 677
Score = 35.4 bits (80), Expect(2) = 3e-54
Identities = 11/21 (52%), Positives = 18/21 (85%)
Frame = +2
Query: 378 LESTLHGIPIVAWPLYADQKM 398
LES +HG+P++ WP++A+Q M
Sbjct: 671 LESVVHGVPLITWPMFAEQGM 733
>TC89198 similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13 {Vigna
angularis}, partial (31%)
Length = 828
Score = 206 bits (523), Expect = 2e-53
Identities = 96/173 (55%), Positives = 127/173 (72%)
Frame = +1
Query: 303 LSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWADQIEILGH 362
LS KFLWV+R P LS DPL+FLP+GFLER K QG V+P WA QI+IL H
Sbjct: 13 LSNHKFLWVVRSPSNTANAAYLSASDVDPLQFLPSGFLERKKEQGMVIPSWAPQIQILRH 192
Query: 363 GAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPNEKGIVG 422
++GGFL HCGWNS LES LHG+P++ WPL+A+Q+ A L S+GLKV LRPK N+ GIV
Sbjct: 193 SSVGGFLTHCGWNSTLESVLHGVPLITWPLFAEQRTNAVLLSEGLKVGLRPKINQNGIVE 372
Query: 423 RGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQLALKWK 475
+ +AE+IK ++ GEEG ++++ MK L++ A A K+DGS T+T++QLALKW+
Sbjct: 373 KVQIAELIKCLMEGEEGGKLRKNMKELKESANSAHKDDGSFTKTLSQLALKWR 531
>TC78509 similar to GP|19911193|dbj|BAB86923. glucosyltransferase-5 {Vigna
angularis}, partial (52%)
Length = 1655
Score = 136 bits (343), Expect(2) = 2e-53
Identities = 71/179 (39%), Positives = 106/179 (58%)
Frame = +2
Query: 300 GLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWADQIEI 359
GLE S +FLW++R + E +LS+ E LP GFLERTK +G VV WA Q I
Sbjct: 959 GLEKSEQRFLWIVRSDMESE---ELSLD-----ELLPEGFLERTKEKGMVVRNWAPQGSI 1114
Query: 360 LGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPNEKG 419
L H ++GGF+ HCGWNS+LE+ G+P++ WPLYA+QKM + KVAL ++ G
Sbjct: 1115 LRHSSVGGFVTHCGWNSVLEAICEGVPMITWPLYAEQKMNRLILVQEWKVALELNESKDG 1294
Query: 420 IVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQLALKWKRSA 478
V + E +K ++ E+GKE+++ + +++ A +A GSS + +L W+ A
Sbjct: 1295 FVSENELGERVKELMESEKGKEVRETILKMKISAKEARGGGGSSLVDLKKLGDSWREHA 1471
Score = 90.9 bits (224), Expect(2) = 2e-53
Identities = 79/307 (25%), Positives = 143/307 (45%), Gaps = 18/307 (5%)
Frame = +3
Query: 11 IVSVPLYS--HTRTILEFCKRLAHLHQD--IHITCINP------TVKSSCNDVKALFENL 60
IV P + H +++E K + H I I + P T+ S + + N
Sbjct: 90 IVLYPAFGSGHLMSMVELGKLILTHHPSFSIKILILTPPNQDTNTINVSTSQYISSVSNK 269
Query: 61 PPNIDSMFLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVD 120
P+I+ ++P ++ PH+ L + ++ +++ L+++ +SN L ++++D
Sbjct: 270 FPSINFHYIPSISFT-FTLPPHLQTLELSPRSNH--HVHHILQSIAKTSN---LKAVMLD 431
Query: 121 VLITQVLPMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISCET--------VEIPGCM 172
L + N L + +Y Y++S A LL L L TF K + +E+PG
Sbjct: 432 FLNYSASQVTNNLEIPTYFYYTSGASLLCLFLNFPTFHKNATIPIKDYNMHTPIELPGLP 611
Query: 173 LIHTTDLPDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTS 232
+ D PD + + S Y+V L+ K +DG+I+N+F +E + +AL+
Sbjct: 612 RLSKEDYPD--EGKDPSSPSYQVLLQSAKSLRESDGIIVNTFDAIEKKAIKALRNGLCVP 785
Query: 233 STSIPHVYPVGPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHD 292
+ P ++ +GP+V SCE +S + WLD Q SV+ +SFGS G +
Sbjct: 786 DGTTPLLFCIGPVVST-SCEEDKS--------GCLSWLDSQPGQSVVLLSFGSLGRFSKA 938
Query: 293 QFNELAL 299
Q N++A+
Sbjct: 939 QINQIAM 959
>TC88760 similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13 {Vigna
angularis}, partial (34%)
Length = 862
Score = 185 bits (469), Expect = 4e-47
Identities = 110/272 (40%), Positives = 153/272 (55%), Gaps = 6/272 (2%)
Frame = +3
Query: 6 TTHIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNID 65
T HIA+V YSH IL+F KRL LH D H+TC PT+ S N K++ + LP NI+
Sbjct: 54 TVHIAVVPGVGYSHLVPILQFSKRLVQLHPDFHVTCFIPTLGSPSNATKSILQTLPSNIN 233
Query: 66 SMFLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVLITQ 125
FLPPVN +D+P+ + + T+ +SLP +++ALK+L S L ++VVD +
Sbjct: 234 HTFLPPVNPNDLPQGTTMESQMFLTLNNSLPYLHDALKSLAIES---PLVALVVDSFAVE 404
Query: 126 VLPMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISC------ETVEIPGCMLIHTTDL 179
VL + ELN+LSYVYF + A L+ +Y D+ SC E ++IPGC+ IH DL
Sbjct: 405 VLNIGKELNMLSYVYFPAAATTLAWSIYLPKLDEETSCEYRDIPEPIKIPGCVPIHGRDL 584
Query: 180 PDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTSIPHV 239
Q+ R S+ YK FL K ADGV +NSF +LE A++E + P V
Sbjct: 585 LSVAQD--RSSQVYKHFLPLFKLLSFADGVFVNSFLELEMGPISAMKE----EGSDNPPV 746
Query: 240 YPVGPLVQEGSCETHESQGNEGETHEYMRWLD 271
YPVGP++Q T S G++ E + WLD
Sbjct: 747 YPVGPIIQ-----TETSSGDDANALECLAWLD 827
>TC82297 similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13 {Vigna
angularis}, partial (13%)
Length = 875
Score = 178 bits (452), Expect(2) = 6e-47
Identities = 102/272 (37%), Positives = 155/272 (56%), Gaps = 6/272 (2%)
Frame = +2
Query: 8 HIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNIDSM 67
HI +V ++H ILEF KRL +LH HITCI P++ S + K+ + LPP I S+
Sbjct: 59 HIVVVPSAGFTHLVPILEFSKRLVNLHPQFHITCIIPSIGSPPSSSKSYLQTLPPTISSI 238
Query: 68 FLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVLITQVL 127
FLPP+N+D +P+ + + + ++ SLP I LK+L CS + + ++V DV VL
Sbjct: 239 FLPPINVDQVPDAKILAVQISLSVKHSLPYIEQELKSL-CSRSKV--VAVVADVFAHDVL 409
Query: 128 PMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISCET------VEIPGCMLIHTTDLPD 181
+A + N+L Y+Y AM+LS Y S D+ +S E+ +++PGC+ DLP
Sbjct: 410 DIAKDFNLLCYIYLPQAAMVLSTYFYSSKLDEILSDESRDPNEPIKVPGCVAFDLKDLPL 589
Query: 182 PVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTSIPHVYP 241
P RFR + Y FLE ++++L DGV +NSF + E + + L+E K P VYP
Sbjct: 590 PF--RFRSNIGYTKFLERAEKYHLFDGVFVNSFLEFEEDAIKGLKEEK-----KKPMVYP 748
Query: 242 VGPLVQEGSCETHESQGNEGETHEYMRWLDKQ 273
VGP++Q+ S G+E E + + WL+KQ
Sbjct: 749 VGPIIQKVSI------GDENEV-KCLTWLEKQ 823
Score = 27.3 bits (59), Expect(2) = 6e-47
Identities = 12/19 (63%), Positives = 14/19 (73%)
Frame = +3
Query: 272 KQEQNSVLYVSFGSAGTLT 290
K SVL+VSFGS GTL+
Sbjct: 816 KNRPKSVLFVSFGSGGTLS 872
>TC87866 similar to GP|19911195|dbj|BAB86924. glucosyltransferase-6 {Vigna
angularis}, partial (12%)
Length = 960
Score = 121 bits (303), Expect(2) = 2e-45
Identities = 83/251 (33%), Positives = 131/251 (52%), Gaps = 11/251 (4%)
Frame = +2
Query: 7 THIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNIDS 66
T+IA+V P SH +EF K+L H + +T I PT+ ++ + LPPNI+
Sbjct: 41 TYIAMVPSPGLSHLIPQVEFAKQLLQQHNEFIVTFIIPTLCPLTPSMQQVLNTLPPNIEF 220
Query: 67 MFLPPVNLDDMPEKPHIPIL-VHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVLITQ 125
+ LP V +PE P + + S+P + +K+L +N L S++ + T
Sbjct: 221 IVLPQVK--HLPEINLEPATQMKLIVKHSIPFLQEEVKSLISKTN---LVSLIFGLFSTD 385
Query: 126 VLPMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISC---------ETVEIPGCML-IH 175
+A + N+ SY++++S A+ LS L D ++S ETV IPGC++ H
Sbjct: 386 AHEVAKQFNLSSYLFYASGALSLSFFLTLPNLDDSVSSEAEFLESAYETVNIPGCVIPFH 565
Query: 176 TTDLPDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTS 235
D+P+ + R + YK+FLE ++ L DG II++F DLEP+ R LQE +
Sbjct: 566 IKDIPEIILCE-RSNVNYKIFLEVCQKLTLVDGFIISTFTDLEPDVIRVLQEKEK----- 727
Query: 236 IPHVYPVGPLV 246
P VYPVGP++
Sbjct: 728 -PCVYPVGPII 757
Score = 79.7 bits (195), Expect(2) = 2e-45
Identities = 37/65 (56%), Positives = 46/65 (69%)
Frame = +3
Query: 254 HESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLELSGVKFLWVIR 313
+ES + +RWL+ Q NSVLYV FGS GT++H+Q NELA GLELSG KFLWV+R
Sbjct: 762 NESNCEDNINSMCLRWLENQPPNSVLYVCFGSGGTVSHEQLNELAFGLELSGKKFLWVVR 941
Query: 314 PPQKL 318
P K+
Sbjct: 942 VPSKV 956
>TC83353 similar to GP|19911195|dbj|BAB86924. glucosyltransferase-6 {Vigna
angularis}, partial (5%)
Length = 789
Score = 179 bits (453), Expect = 3e-45
Identities = 102/257 (39%), Positives = 148/257 (56%), Gaps = 6/257 (2%)
Frame = +1
Query: 2 EVTTTTHIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLP 61
E T+HI + S+PL+SH +++EFCKRL +H ITCI PT+ + L E+LP
Sbjct: 34 ETKPTSHIVVTSIPLFSHESSVIEFCKRLIQVHNHFQITCIFPTIDAPIPATLKLLESLP 213
Query: 62 PNIDSMFLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDV 121
I FLPP+ D+P++ + + + +T S+PS +L L CS+++ + +IVVD
Sbjct: 214 STIHCTFLPPIKKQDLPQE--VTMQLELGVTKSMPSFRESLSLL-CSTSTTPVVAIVVDP 384
Query: 122 LITQVLPMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTIS------CETVEIPGCMLIH 175
Q L +A E N+LS++YF +AM SL L+ D+ +S E +EIPGC I
Sbjct: 385 FANQALEIAKEFNILSFMYFPVSAMTTSLHLHLPILDEQVSGEYMDHVEPIEIPGCTPIR 564
Query: 176 TTDLPDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTS 235
DLP R S Y+ L KRF LADGV+INSF ++E T RAL E + +++
Sbjct: 565 GQDLPRTFFED-RSSIAYETILRQTKRFSLADGVLINSFSEMEESTVRALMEKEQSNNKQ 741
Query: 236 IPHVYPVGPLVQEGSCE 252
+ VY VGP++Q GS E
Sbjct: 742 L--VYLVGPIIQTGSNE 786
>CB892343 similar to GP|19911195|dbj glucosyltransferase-6 {Vigna angularis},
partial (5%)
Length = 811
Score = 176 bits (446), Expect = 2e-44
Identities = 110/276 (39%), Positives = 153/276 (54%), Gaps = 8/276 (2%)
Frame = +1
Query: 8 HIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNIDSM 67
HIA+VS+PL+SH +I+EFCKRL HLH IHITCI T+ + L E+LP +I+
Sbjct: 19 HIAVVSIPLFSHQSSIIEFCKRLIHLHHHIHITCIFSTIDAPIPATLKLLESLPSSINCT 198
Query: 68 FLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVLITQVL 127
FLPP+N D+P + + T S+PS +L +L SS S + ++VVD +Q L
Sbjct: 199 FLPPINKQDLPR--DFVLEIELTTAQSMPSFRKSLLSLCSSSTSSPVVALVVDPYASQAL 372
Query: 128 PMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISCE------TVEIPGCMLIHTTDLPD 181
+A + N+LS+VYF +AM S LY + +SCE ++ PGC+ I + DLP
Sbjct: 373 EIAKD*NILSFVYFPLSAMTTSFNLYFPALHEQVSCEYKDHTDLIQFPGCLPICSQDLP- 549
Query: 182 PVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQE--NKGTSSTSIPHV 239
P R S Y FL +K LA G ++NSF +E T RALQE NK T V
Sbjct: 550 PEFFHDRSSVRYSFFLLHSKNLSLAHGFLVNSFSKMEASTGRALQEEHNKTTKL-----V 714
Query: 240 YPVGPLVQEGSCETHESQGNEGETHEYMRWLDKQEQ 275
Y VG ++Q GS + ES G+ ++WL+ Q
Sbjct: 715 YMVGTIIQSGSNCSDESNGS-----ICLKWLEINTQ 807
>TC79143 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (38%)
Length = 698
Score = 166 bits (420), Expect = 2e-41
Identities = 88/233 (37%), Positives = 139/233 (58%), Gaps = 6/233 (2%)
Frame = +3
Query: 1 MEVTTTTHIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENL 60
+ + T+HIA++S P +SH I+EF KRL H + H+TCI P++ S + K+ E +
Sbjct: 12 LTMAKTSHIAVISSPGFSHIAPIVEFSKRLVTNHPNFHVTCIIPSLGSLQDSSKSYLETV 191
Query: 61 PPNIDSMFLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVD 120
PPNI+ +FLPP+N D+P+ + IL+ T+T SLPSI+ ALK++ ++ L +I+ D
Sbjct: 192 PPNINLVFLPPINKQDLPQGVYPGILIQLTVTRSLPSIHQALKSI---NSKAPLVAIIAD 362
Query: 121 VLITQVLPMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISC------ETVEIPGCMLI 174
+ L A E N LSYVYF +A +LS L+ D+ +SC E +++ GC+ I
Sbjct: 363 NFAWEALDFAKEFNSLSYVYFPCSAFVLSFYLHWPKLDEEVSCKYKDLQEPIKLQGCVPI 542
Query: 175 HTTDLPDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQE 227
+ DLP ++ R + YK++L+ K DG++ NSFF LE +AL++
Sbjct: 543 NGIDLPTVTKD--RSGQAYKMYLQRAKDMCFVDGILFNSFFALESSAIKALEQ 695
>TC87302 similar to GP|19911199|dbj|BAB86926. glucosyltransferase-8 {Vigna
angularis}, partial (53%)
Length = 1787
Score = 164 bits (416), Expect = 5e-41
Identities = 98/285 (34%), Positives = 151/285 (52%), Gaps = 8/285 (2%)
Frame = +1
Query: 194 KVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTSIPHVYPVGPLVQEGSCET 253
K+ E N+ + GVI N+F++LEP + G + + V +E +C
Sbjct: 622 KLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHLGPVSLCNRDTEEKACRG 801
Query: 254 HESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLELSGVKFLWVIR 313
E+ +E HE ++WL +E NSV+YV FGS + Q E+A+GLE S V F+WV+R
Sbjct: 802 REASIDE---HECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVR 972
Query: 314 PPQKLEMIGDLSVGHEDPLEFLPNGFLERTKG--QGFVVPYWADQIEILGHGAIGGFLCH 371
K E + LE+LP GF ER +G +G ++ WA Q+ IL H ++GGF+ H
Sbjct: 973 KSAKSE---------GENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTH 1125
Query: 372 CGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPNEKGIVGRG------V 425
CGWNS LE G+P+V WP+Y +Q A SD +K+ + +G G V
Sbjct: 1126CGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEPVKKDV 1305
Query: 426 VAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQL 470
+ + ++ I+VG+E +E++ R K +A A++ GSS + L
Sbjct: 1306IEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNL 1440
>TC78077 similar to GP|4115536|dbj|BAA36411.1 UDP-glycose:flavonoid
glycosyltransferase {Vigna mungo}, partial (66%)
Length = 865
Score = 155 bits (391), Expect = 4e-38
Identities = 79/201 (39%), Positives = 122/201 (60%)
Frame = +1
Query: 278 VLYVSFGSAGTLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPN 337
V+ +SFGS G + Q NE+A+GLE S +FLWV+R + LS+ E P
Sbjct: 1 VVLLSFGSMGRFSRAQLNEIAIGLEKSEQRFLWVVRSEPDSDK---LSLD-----ELFPE 156
Query: 338 GFLERTKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQK 397
GFLERTK +G VV WA Q+ IL H ++GGF+ HCGWNS+LE+ G+P++AWPL+A+Q+
Sbjct: 157 GFLERTKDKGMVVRNWAPQVAILSHNSVGGFVTHCGWNSVLEAICEGVPMIAWPLFAEQR 336
Query: 398 MIAALFSDGLKVALRPKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDAL 457
+ + D +KVAL+ +E V + E +K ++ + GK+I++R+ +++ A +A
Sbjct: 337 LNRLVLVDEMKVALKVNQSENRFVSGTELGERVKELMESDRGKDIKERILKMKISAKEAR 516
Query: 458 KEDGSSTRTITQLALKWKRSA 478
GSS + +L W+ A
Sbjct: 517 GGGGSSLVDLKKLGDSWREHA 579
>TC85899 weakly similar to GP|7385017|gb|AAF61647.1| UDP-glucose:salicylic
acid glucosyltransferase {Nicotiana tabacum}, partial
(47%)
Length = 1617
Score = 150 bits (378), Expect = 1e-36
Identities = 129/473 (27%), Positives = 215/473 (45%), Gaps = 11/473 (2%)
Frame = +2
Query: 8 HIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPT----VKSSCNDVKALFENLPPN 63
H I+ P H +++F KRL + + + IT I T V S+ N E++
Sbjct: 44 HCLILPYPAQGHMNPMIQFSKRL--IEKGVKITLITVTSFWKVISNKNLTSIDVESISDG 217
Query: 64 IDSM-FLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVL 122
D L +L+D E + +L + + L SS+ ++ D
Sbjct: 218 YDEGGLLAAESLEDYKET------FWKVGSQTLSELLHKL-----SSSENPPNCVIFDAF 364
Query: 123 ITQVLPMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISCETVE----IPGCMLIHTTD 178
+ VL + ++ +F+ + + S+ Y T +K I + +PG + D
Sbjct: 365 LPWVLDVGKSFGLVGVAFFTQSCSVNSV--YYHTHEKLIELPLTQSEYLLPGLPKLAPGD 538
Query: 179 LPDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQE--NKGTSSTSI 236
LP + + + + AD ++ NS ++LEPE L + T S+
Sbjct: 539 LPSFLYKYGSYPGYFDIVVNQFANIGKADWILANSIYELEPEVVDWLVKIWPLKTIGPSV 718
Query: 237 PHVYPVGPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNE 296
P + L + S N T ++WL+ + + SV+Y SFGS L+ +Q E
Sbjct: 719 PSMLLDKRLKDDKEYGVSLSDPN---TEFCIKWLNDKPKGSVVYASFGSMAGLSEEQTQE 889
Query: 297 LALGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWADQ 356
LALGL+ S FLWV+R + + LP GF+E +K +G +V W Q
Sbjct: 890 LALGLKDSESYFLWVVRECDQSK---------------LPKGFVESSK-KGLIVT-WCPQ 1018
Query: 357 IEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPN 416
+ +L H A+G F+ HCGWNS LE+ G+P++A PL+ DQ A L +D K+ +R +
Sbjct: 1019LLVLTHEALGCFVTHCGWNSTLEALSIGVPLIAMPLWTDQVTNAKLIADVWKMGVRAVAD 1198
Query: 417 EKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQ 469
EK IV + IK I+ E+G EI++ + +++A ++ E G S + I +
Sbjct: 1199EKEIVRSETIKNCIKEIIETEKGNEIKKNALKWKNLAKSSVDEGGRSDKNIEE 1357
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,825,986
Number of Sequences: 36976
Number of extensions: 240049
Number of successful extensions: 1615
Number of sequences better than 10.0: 149
Number of HSP's better than 10.0 without gapping: 1475
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1480
length of query: 479
length of database: 9,014,727
effective HSP length: 100
effective length of query: 379
effective length of database: 5,317,127
effective search space: 2015191133
effective search space used: 2015191133
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0135.13


