

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
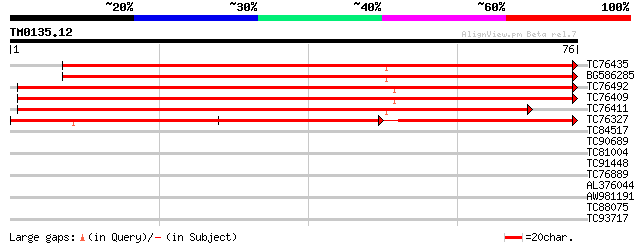
Query= TM0135.12
(76 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76435 homologue to GP|13359451|dbj|BAB33421. putative senescen... 116 1e-27
BG586285 homologue to GP|13359451|db putative senescence-associa... 111 6e-26
TC76492 similar to GP|13359451|dbj|BAB33421. putative senescence... 84 8e-18
TC76409 similar to GP|13359451|dbj|BAB33421. putative senescence... 79 4e-16
TC76411 weakly similar to GP|13359451|dbj|BAB33421. putative sen... 67 1e-12
TC76327 similar to GP|13359451|dbj|BAB33421. putative senescence... 52 3e-08
TC84517 27 1.1
TC90689 similar to GP|4099092|gb|AAD09232.1| unknown {Arabidopsi... 27 1.5
TC81004 weakly similar to SP|Q39529|LEC2_CLALU Agglutinin II pre... 26 2.5
TC91448 weakly similar to PIR|C86432|C86432 protein T5I8.13 [imp... 26 3.3
TC76889 weakly similar to PIR|F81516|F81516 hypothetical protein... 25 4.2
AL376044 similar to GP|18656155|db ryanodine receptor {Hemicentr... 24 9.5
AW981191 24 9.5
TC88075 homologue to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia ... 24 9.5
TC93717 similar to GP|7262671|gb|AAF43929.1| EST gb|AI997595 com... 24 9.5
>TC76435 homologue to GP|13359451|dbj|BAB33421. putative
senescence-associated protein {Pisum sativum}, complete
Length = 1389
Score = 116 bits (291), Expect = 1e-27
Identities = 56/71 (78%), Positives = 59/71 (82%), Gaps = 2/71 (2%)
Frame = +1
Query: 8 SPNSELEALSHNPTHGSFAPLTSQTSVMTNCANQRFLFN*VE--LRHFHQ*GKTNMSHDG 65
SP+S+LEA SHNPTHGSFAPL Q S MTNCANQRFL *VE LRH HQ*GKTN+SHDG
Sbjct: 391 SPDSDLEAFSHNPTHGSFAPLAFQPSAMTNCANQRFLSY*VELLLRHCHQ*GKTNLSHDG 570
Query: 66 LIPTHVPYWWV 76
L P HVPYWWV
Sbjct: 571 LNPAHVPYWWV 603
>BG586285 homologue to GP|13359451|db putative senescence-associated protein
{Pisum sativum}, partial (45%)
Length = 704
Score = 111 bits (277), Expect = 6e-26
Identities = 54/71 (76%), Positives = 57/71 (80%), Gaps = 2/71 (2%)
Frame = +3
Query: 8 SPNSELEALSHNPTHGSFAPLTSQTSVMTNCANQRFLFN*VE--LRHFHQ*GKTNMSHDG 65
SP+S+LEA SHNP HGSFAPL Q S MTNCANQRFL *VE LR HQ*GKTN+SHDG
Sbjct: 297 SPDSDLEAFSHNPAHGSFAPLAFQPSAMTNCANQRFLSY*VELLLRRGHQ*GKTNLSHDG 476
Query: 66 LIPTHVPYWWV 76
L P HVPYWWV
Sbjct: 477 LNPAHVPYWWV 509
>TC76492 similar to GP|13359451|dbj|BAB33421. putative senescence-associated
protein {Pisum sativum}, partial (35%)
Length = 829
Score = 84.3 bits (207), Expect = 8e-18
Identities = 43/77 (55%), Positives = 51/77 (65%), Gaps = 2/77 (2%)
Frame = +1
Query: 2 EYTHRYSPNSELEALSHNPTHGSFAPLTSQTSVMTNCANQRFLFN*VEL--RHFHQ*GKT 59
E + R +S+LEA SH P GS A L +T+ TN N+RFL * +L R HQ*GKT
Sbjct: 115 EVSSRSGMDSDLEAFSHYPADGSVAALPGRTAARTNYLNERFLSY*AQLPLRRGHQ*GKT 294
Query: 60 NMSHDGLIPTHVPYWWV 76
N+SHDGL P HVPYWWV
Sbjct: 295 NLSHDGLNPAHVPYWWV 345
>TC76409 similar to GP|13359451|dbj|BAB33421. putative senescence-associated
protein {Pisum sativum}, partial (40%)
Length = 614
Score = 78.6 bits (192), Expect = 4e-16
Identities = 42/77 (54%), Positives = 50/77 (64%), Gaps = 2/77 (2%)
Frame = -3
Query: 2 EYTHRYSPNSELEALSHNPTHGSFAPLTSQTSVMTNCANQRFLFN*VEL--RHFHQ*GKT 59
E + R +S+LEA SH P GS A L +T+ TN N+RFL * +L R HQ*GKT
Sbjct: 402 EVSSRSGMDSDLEAFSHYPADGSVAALPGRTAARTNYLNERFLSY*AQLPLRRGHQ*GKT 223
Query: 60 NMSHDGLIPTHVPYWWV 76
N+SHDGL P HVPY WV
Sbjct: 222 NLSHDGLNPAHVPY*WV 172
>TC76411 weakly similar to GP|13359451|dbj|BAB33421. putative
senescence-associated protein {Pisum sativum}, partial
(22%)
Length = 592
Score = 67.4 bits (163), Expect = 1e-12
Identities = 37/71 (52%), Positives = 45/71 (63%), Gaps = 2/71 (2%)
Frame = +2
Query: 2 EYTHRYSPNSELEALSHNPTHGSFAPLTSQTSVMTNCANQRFLFN*VE--LRHFHQ*GKT 59
E + R +S+LEA SH P GS A L +T+ TN N+RFL * + LR HQ*GKT
Sbjct: 380 EVSSRSGMDSDLEAFSHYPADGSVAALPGRTAARTNYLNERFLSY*AQLPLRRGHQ*GKT 559
Query: 60 NMSHDGLIPTH 70
N+SHDGL P H
Sbjct: 560 NLSHDGLNPAH 592
>TC76327 similar to GP|13359451|dbj|BAB33421. putative
senescence-associated protein {Pisum sativum}, partial
(64%)
Length = 868
Score = 52.4 bits (124), Expect = 3e-08
Identities = 30/51 (58%), Positives = 34/51 (65%), Gaps = 1/51 (1%)
Frame = +2
Query: 1 NEYTHRY-SPNSELEALSHNPTHGSFAPLTSQTSVMTNCANQRFLFN*VEL 50
N+ HR+ S +S LEA SHNPT GSFA L Q + TN NQRFL *VEL
Sbjct: 5 NKSHHRFFSMDSGLEAFSHNPTDGSFASLAFQLNAFTNYLNQRFLSY*VEL 157
Score = 47.0 bits (110), Expect = 1e-06
Identities = 23/48 (47%), Positives = 31/48 (63%)
Frame = +3
Query: 29 TSQTSVMTNCANQRFLFN*VELRHFHQ*GKTNMSHDGLIPTHVPYWWV 76
+S+T + C ++ +F+ HQ*GKTN+SHDGL P HVPY WV
Sbjct: 135 SSRTELNYYCDDKMKIFS------IHQ*GKTNLSHDGLNPAHVPY*WV 260
>TC84517
Length = 455
Score = 27.3 bits (59), Expect = 1.1
Identities = 15/37 (40%), Positives = 18/37 (48%)
Frame = +1
Query: 1 NEYTHRYSPNSELEALSHNPTHGSFAPLTSQTSVMTN 37
N + PNS +LSH P H P T+QT TN
Sbjct: 310 NHFARVKVPNSSNPSLSHRPMH---LPKTTQTCSCTN 411
>TC90689 similar to GP|4099092|gb|AAD09232.1| unknown {Arabidopsis
thaliana}, partial (46%)
Length = 830
Score = 26.9 bits (58), Expect = 1.5
Identities = 15/35 (42%), Positives = 19/35 (53%)
Frame = +1
Query: 5 HRYSPNSELEALSHNPTHGSFAPLTSQTSVMTNCA 39
HR SP+S L L H P+ S LTS +V + A
Sbjct: 223 HRLSPSSTLTQLIHPPSALSAHKLTSSLTVSAHSA 327
>TC81004 weakly similar to SP|Q39529|LEC2_CLALU Agglutinin II precursor
(ClAII) (LecClAII). [Yellow wood] {Cladrastis lutea},
partial (24%)
Length = 610
Score = 26.2 bits (56), Expect = 2.5
Identities = 16/46 (34%), Positives = 23/46 (49%), Gaps = 1/46 (2%)
Frame = -2
Query: 32 TSVMTNCANQRFLFN*VELRHFHQ*GKTNMSHDGL-IPTHVPYWWV 76
T ++TNC+N R *+ L + DG+ I TH+ WWV
Sbjct: 378 TIIITNCSNHR*---*IALNTINSPSLRCFGTDGICINTHMRIWWV 250
>TC91448 weakly similar to PIR|C86432|C86432 protein T5I8.13 [imported] -
Arabidopsis thaliana, partial (8%)
Length = 682
Score = 25.8 bits (55), Expect = 3.3
Identities = 10/29 (34%), Positives = 16/29 (54%)
Frame = +1
Query: 17 SHNPTHGSFAPLTSQTSVMTNCANQRFLF 45
SH P HG+ +T +T + + N RF +
Sbjct: 4 SHAPFHGTLFTVTPKTKTLHSSQNMRFRY 90
>TC76889 weakly similar to PIR|F81516|F81516 hypothetical protein CP0987
[imported] - Chlamydophila pneumoniae (strain AR39),
partial (62%)
Length = 2758
Score = 25.4 bits (54), Expect = 4.2
Identities = 10/14 (71%), Positives = 11/14 (78%)
Frame = +2
Query: 59 TNMSHDGLIPTHVP 72
T +SHD L PTHVP
Sbjct: 254 TELSHDVLNPTHVP 295
>AL376044 similar to GP|18656155|db ryanodine receptor {Hemicentrotus
pulcherrimus}, partial (0%)
Length = 525
Score = 24.3 bits (51), Expect = 9.5
Identities = 9/17 (52%), Positives = 10/17 (57%)
Frame = +1
Query: 54 HQ*GKTNMSHDGLIPTH 70
HQ GK N H G +P H
Sbjct: 379 HQGGKPNFQHKGGLPNH 429
>AW981191
Length = 656
Score = 24.3 bits (51), Expect = 9.5
Identities = 10/26 (38%), Positives = 15/26 (57%)
Frame = +3
Query: 9 PNSELEALSHNPTHGSFAPLTSQTSV 34
P+ L+ + H+P+ SFA LT V
Sbjct: 549 PSVALQKMQHHPSSSSFADLTDSEIV 626
>TC88075 homologue to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia x
hybrida}, partial (38%)
Length = 674
Score = 24.3 bits (51), Expect = 9.5
Identities = 9/17 (52%), Positives = 12/17 (69%)
Frame = +2
Query: 5 HRYSPNSELEALSHNPT 21
HR+S NS + S+NPT
Sbjct: 68 HRFSNNSNVTIFSYNPT 118
>TC93717 similar to GP|7262671|gb|AAF43929.1| EST gb|AI997595 comes from
this gene. {Arabidopsis thaliana}, partial (51%)
Length = 699
Score = 24.3 bits (51), Expect = 9.5
Identities = 9/17 (52%), Positives = 12/17 (69%)
Frame = -1
Query: 17 SHNPTHGSFAPLTSQTS 33
+H P+HG F +TSQ S
Sbjct: 477 NHRPSHGWFYKMTSQAS 427
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.332 0.139 0.486
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,004,907
Number of Sequences: 36976
Number of extensions: 38419
Number of successful extensions: 292
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 286
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 287
length of query: 76
length of database: 9,014,727
effective HSP length: 52
effective length of query: 24
effective length of database: 7,091,975
effective search space: 170207400
effective search space used: 170207400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0135.12


