

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0134.21
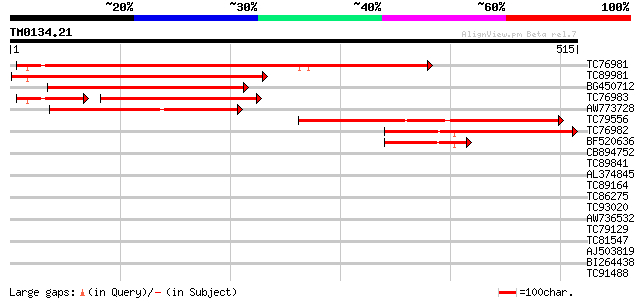
(515 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76981 similar to PIR|T12198|T12198 sucrose transport protein -... 514 e-146
TC89981 similar to GP|9957218|gb|AAG09270.1| sucrose transporter... 293 1e-79
BG450712 similar to GP|17645762|em unnamed protein product {Glyc... 293 1e-79
TC76983 similar to PIR|T12198|T12198 sucrose transport protein -... 229 3e-77
AW773728 similar to GP|10998390|gb sucrose transporter SUT4 {Sol... 245 3e-65
TC79556 similar to GP|5882292|gb|AAD55269.1| sucrose transporter... 223 1e-58
TC76982 similar to PIR|T12198|T12198 sucrose transport protein -... 223 1e-58
BF520636 homologue to GP|5230818|gb|A sucrose transport protein ... 84 1e-16
CB894752 34 0.12
TC89841 similar to PIR|T02307|T02307 probable membrane transport... 33 0.20
AL374845 similar to GP|12247468|gb| PnFL-1 {Ipomoea nil}, partia... 33 0.34
TC89164 weakly similar to GP|10177340|dbj|BAB10596. transporter-... 32 0.45
TC86275 similar to PIR|E96603|E96603 unknown protein F14G9.26 [i... 32 0.59
TC93020 similar to GP|6635840|gb|AAF20003.1| 1-acyl-sn-glycerol-... 31 1.3
AW736532 similar to GP|15983497|gb| AT5g38560/MBB18_10 {Arabidop... 30 1.7
TC79129 similar to PIR|A96725|A96725 hypothetical protein F20P5.... 30 2.2
TC81547 similar to GP|13877739|gb|AAK43947.1 unknown protein {Ar... 30 2.9
AJ503819 30 2.9
BI264438 similar to GP|21427467|gb actin-related protein 6 {Arab... 30 2.9
TC91488 similar to GP|11078533|gb|AAG29095.1 Glu-tRNA(Gln) amido... 29 3.8
>TC76981 similar to PIR|T12198|T12198 sucrose transport protein - fava bean,
partial (71%)
Length = 1224
Score = 514 bits (1325), Expect = e-146
Identities = 257/392 (65%), Positives = 304/392 (76%), Gaps = 14/392 (3%)
Frame = +2
Query: 7 PPNSTTQN---NNSIAPSSFQVEPAQHAAGPSPLAKMIAVASIAAGVQFGWALQLSLLTP 63
P +ST QN NN++ S VE PSPL K+I VASIAAGVQFGWALQLSLLTP
Sbjct: 44 PFSSTKQNHNNNNTLTKPSLHVESPP--LEPSPLRKIIVVASIAAGVQFGWALQLSLLTP 217
Query: 64 YVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGAIAVAIAVF 123
YVQLLG+PH W+++IWLCGPISG++VQPIVGY SDRCTSRFGRRRPFI AG+ AVAIAVF
Sbjct: 218 YVQLLGIPHTWAAYIWLCGPISGMLVQPIVGYHSDRCTSRFGRRRPFIAAGSFAVAIAVF 397
Query: 124 LIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWILDVANNMLQGPCRAFLGDLSTGHHSR 183
LIG+AAD+G+S GDDLSKK RPRA+ FV+GFWILDVANNMLQGPCRA LGDL G+H +
Sbjct: 398 LIGYAADLGHSFGDDLSKKVRPRAIGIFVVGFWILDVANNMLQGPCRALLGDLCAGNHQK 577
Query: 184 IRTANTIFSFFMGVGNVLGYLAGSYGGLHKIFPFTETKACDVFCANLKSCFFFSITLLLV 243
R AN FSFFM VGN+LGY AG+Y L +FPFT+TKACD++CANLKSCFF SI LL
Sbjct: 578 TRNANAFFSFFMAVGNILGYAAGAYSKLFHVFPFTKTKACDIYCANLKSCFFLSIALLTA 757
Query: 244 LSGFALFYVHDPPIGSRR-------EEDDKAPKN---VFVELFGAFKELKKPMLMLMLVT 293
++ AL YV + P+ + +ED K+ F EL GAF+ELK+PM +L+LVT
Sbjct: 758 VATAALIYVKEIPLSPEKVTGNGVTDEDGNVTKSSNPCFGELSGAFRELKRPMWILLLVT 937
Query: 294 SLNWIAWFPYVLYDTDWMGLEVYGGKLG-SKAYDAGVRAGALGLVLNSVVLGLMSLAVEP 352
LNWIAWFP++L+DTDWMG EVYGG +G AYD GVRAGALGL+LNSVVLG SL V+
Sbjct: 938 CLNWIAWFPFLLFDTDWMGKEVYGGTVGEGHAYDKGVRAGALGLMLNSVVLGATSLXVDV 1117
Query: 353 LGRYLGGVKRLWAIVNIILAVCMAMTMLITKV 384
L R +GGVKRLW IVN +LA+C+AMT+ TK+
Sbjct: 1118LARGVGGVKRLWGIVNFLLAICLAMTVXATKL 1213
>TC89981 similar to GP|9957218|gb|AAG09270.1| sucrose transporter
{Lycopersicon esculentum}, partial (39%)
Length = 717
Score = 293 bits (750), Expect = 1e-79
Identities = 140/237 (59%), Positives = 170/237 (71%), Gaps = 4/237 (1%)
Frame = +3
Query: 2 APPPPPPNSTTQN----NNSIAPSSFQVEPAQHAAGPSPLAKMIAVASIAAGVQFGWALQ 57
A P PN TT N + S+ P Q +PL +++ VAS+A+G+QFGWALQ
Sbjct: 6 ASASPMPNPTTTNPHRSRTRSSTSTSTSRPVQPVQPRTPLRQLLRVASVASGIQFGWALQ 185
Query: 58 LSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGAIA 117
LSLLTPYVQ LG+PHKW+S IWLCGP+SGL VQP+VG+ SDRC+SRFGRRRPFI GA +
Sbjct: 186 LSLLTPYVQQLGIPHKWASIIWLCGPVSGLFVQPLVGHLSDRCSSRFGRRRPFILVGAAS 365
Query: 118 VAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWILDVANNMLQGPCRAFLGDLS 177
+ +AV +IG+AADIGY +GDD+++ RP A+ FVIGFWILDVANN+ QGPCRA L DL+
Sbjct: 366 IVVAVVIIGYAADIGYLIGDDITQNYRPFAIVVFVIGFWILDVANNVTQGPCRALLADLT 545
Query: 178 TGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHKIFPFTETKACDVFCANLKSCF 234
R R AN FS FM VGN+LGY GSY G +KIF FT T AC + CAN KS F
Sbjct: 546 CNDARRTRVANAYFSLFMAVGNILGYATGSYSGWYKIFTFTLTPACSIXCANXKSAF 716
>BG450712 similar to GP|17645762|em unnamed protein product {Glycine max},
partial (35%)
Length = 555
Score = 293 bits (750), Expect = 1e-79
Identities = 140/183 (76%), Positives = 158/183 (85%)
Frame = +2
Query: 35 SPLAKMIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVG 94
+PL KM AVASIAAG+QFGWALQLSLLTPYVQLLGVPH+W++ IWLCGPISG+++QP+VG
Sbjct: 5 TPLRKMAAVASIAAGIQFGWALQLSLLTPYVQLLGVPHQWAANIWLCGPISGMIIQPLVG 184
Query: 95 YSSDRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVIG 154
Y SDR SRFGRRRPFIF GAIAVAIAVFLIG+AAD+G+SMGDDL+KKTRPRAV FV G
Sbjct: 185 YYSDRSHSRFGRRRPFIFFGAIAVAIAVFLIGYAADLGHSMGDDLTKKTRPRAVVIFVFG 364
Query: 155 FWILDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHKI 214
FWILDVANNMLQGPCRAF+GDL+ G H +R N +FSFFM GNVLGY AGSY L+
Sbjct: 365 FWILDVANNMLQGPCRAFIGDLAGGDHRXMRIGNGMFSFFMAXGNVLGYAAGSYXKLYTK 544
Query: 215 FPF 217
PF
Sbjct: 545 XPF 553
>TC76983 similar to PIR|T12198|T12198 sucrose transport protein - fava bean,
partial (40%)
Length = 674
Score = 229 bits (585), Expect(2) = 3e-77
Identities = 107/146 (73%), Positives = 123/146 (83%)
Frame = +2
Query: 83 PISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKK 142
PISG++VQPIVGY SDRCTSRFGRRRPFI AG+ AVAIAVFLIG+AAD+G+S GDDLSKK
Sbjct: 227 PISGMLVQPIVGYHSDRCTSRFGRRRPFIAAGSFAVAIAVFLIGYAADLGHSFGDDLSKK 406
Query: 143 TRPRAVAFFVIGFWILDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLG 202
RPRA+ FV+GFWILDVANNMLQGPCRA LGDL G+H + R AN FSFFM VGN+LG
Sbjct: 407 VRPRAIGIFVVGFWILDVANNMLQGPCRALLGDLCAGNHQKTRNANAFFSFFMAVGNILG 586
Query: 203 YLAGSYGGLHKIFPFTETKACDVFCA 228
Y AG+Y L +FPFT+TKA D++ A
Sbjct: 587 YAAGAYSKLFHVFPFTKTKAXDIYXA 664
Score = 77.8 bits (190), Expect(2) = 3e-77
Identities = 44/68 (64%), Positives = 49/68 (71%), Gaps = 3/68 (4%)
Frame = +3
Query: 7 PPNSTTQN---NNSIAPSSFQVEPAQHAAGPSPLAKMIAVASIAAGVQFGWALQLSLLTP 63
P +ST QN NN++ S VE PSPL K+I VASIAAGVQFGWALQLSLLTP
Sbjct: 30 PFSSTKQNHNNNNTLTKPSLHVESPP--LEPSPLRKIIVVASIAAGVQFGWALQLSLLTP 203
Query: 64 YVQLLGVP 71
YVQLLG+P
Sbjct: 204YVQLLGIP 227
>AW773728 similar to GP|10998390|gb sucrose transporter SUT4 {Solanum
tuberosum}, partial (34%)
Length = 620
Score = 245 bits (626), Expect = 3e-65
Identities = 114/175 (65%), Positives = 137/175 (78%)
Frame = +2
Query: 37 LAKMIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYS 96
L K++ VAS+A G+QFGWALQLSLLTPYVQ LG+PH W+S IWLCGP+SGL+VQP+VG+
Sbjct: 92 LTKLLRVASVAGGIQFGWALQLSLLTPYVQQLGIPHAWASIIWLCGPLSGLLVQPLVGHL 271
Query: 97 SDRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFW 156
SDRCTSRFGRRRPFI GA+++ I+V +IG AAD+G+ GD +K R AVAFFV GFW
Sbjct: 272 SDRCTSRFGRRRPFILGGAVSIVISVLIIGHAADLGWKFGD--TKNHRHSAVAFFVFGFW 445
Query: 157 ILDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGL 211
ILDVANN+ QGPCRA LGDL+ H R AN FS FM +GN+LG+ GSY L
Sbjct: 446 ILDVANNVTQGPCRALLGDLTGKDHRMTRVANAYFSLFMAIGNILGFATGSYSWL 610
>TC79556 similar to GP|5882292|gb|AAD55269.1| sucrose transporter {Vitis
vinifera}, partial (45%)
Length = 1209
Score = 223 bits (568), Expect = 1e-58
Identities = 118/242 (48%), Positives = 155/242 (63%), Gaps = 1/242 (0%)
Frame = +3
Query: 263 EDDKAPKNVFVELFGAFKELKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGKL-G 321
E A + ELFG FK P+ +++ VT+L WI WFP+ L+DTDWMG E+YGG G
Sbjct: 21 ESGSAEEAFMWELFGTFKYFSMPVWIVLSVTALTWIGWFPFNLFDTDWMGREIYGGDPEG 200
Query: 322 SKAYDAGVRAGALGLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLI 381
YD GVR GALGL+LNSVVL + SL +E L R G +W I NI +A+C +++
Sbjct: 201 GLIYDTGVRMGALGLLLNSVVLAVTSLLMERLCRKRGA-GFVWGISNIFMAICFIAMLVL 377
Query: 382 TKVAEHDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQ 441
T A VS G P G+ AL F +LG P+AITYSVP+AL S + G GQ
Sbjct: 378 TYAANSIGYVSKGQP---PPTGIVIAALAIFTILGFPMAITYSVPYALISTHIEPLGLGQ 548
Query: 442 GLSLGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLLPSPKP 501
GLS+GVLN+AIV+PQ++VS +GPWD LFGGGN PAF V AV A +S +LA++ +P +
Sbjct: 549 GLSMGVLNLAIVVPQIVVSLGSGPWDQLFGGGNSPAFAVAAVAALLSGLLALLAIPRTRT 728
Query: 502 EE 503
++
Sbjct: 729 QK 734
>TC76982 similar to PIR|T12198|T12198 sucrose transport protein - fava bean,
partial (40%)
Length = 1085
Score = 223 bits (568), Expect = 1e-58
Identities = 116/178 (65%), Positives = 140/178 (78%), Gaps = 3/178 (1%)
Frame = +2
Query: 341 VVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGGATIGRP 400
VVLG SL V+ L R +GGVKRLW IVN LA+C+AMT+L+TK+A+H R V A+ P
Sbjct: 140 VVLGATSLGVDVLARGVGGVKRLWGIVNFPLAICLAMTVLVTKLAQHSR-VYADASHTDP 316
Query: 401 TP---GVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQGLSLGVLNVAIVIPQM 457
P G+ AGAL F+VLGIPLAITYS+PFALASI+SSS+GAGQGLSLGVLN+AIVIPQM
Sbjct: 317 LPPSGGITAGALALFSVLGIPLAITYSIPFALASIFSSSSGAGQGLSLGVLNLAIVIPQM 496
Query: 458 IVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLLPSPKPEEMAKASISASGFH 515
IVS L+GPWD+LFGGGNLPAF+VGAV A S +L++VLLPSP P+ + + GFH
Sbjct: 497 IVSVLSGPWDALFGGGNLPAFVVGAVAALASGILSVVLLPSPPPDLAKSVTATGGGFH 670
>BF520636 homologue to GP|5230818|gb|A sucrose transport protein SUT1 {Pisum
sativum}, partial (16%)
Length = 334
Score = 84.3 bits (207), Expect = 1e-16
Identities = 47/82 (57%), Positives = 58/82 (70%), Gaps = 3/82 (3%)
Frame = +1
Query: 341 VVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGGATIGRP 400
VVLG SL V+ L R +GGVKRLW IVN LA+C+AMT+L+TK+A+H RV A+ P
Sbjct: 67 VVLGATSLGVDVLARGVGGVKRLWGIVNFPLAICLAMTVLVTKLAQHS-RVYADASHTDP 243
Query: 401 TP---GVKAGALMFFAVLGIPL 419
P G+ AGAL F+VLGIPL
Sbjct: 244 FPPSGGITAGALALFSVLGIPL 309
>CB894752
Length = 152
Score = 34.3 bits (77), Expect = 0.12
Identities = 19/49 (38%), Positives = 26/49 (52%)
Frame = +1
Query: 195 MGVGNVLGYLAGSYGGLHKIFPFTETKACDVFCANLKSCFFFSITLLLV 243
M VG ++GY GS+ G KI T A + AN KS FF + ++V
Sbjct: 1 MAVGTIVGYALGSFSGWDKIRTPILTVASFISGANHKSAFFPDVAAIVV 147
>TC89841 similar to PIR|T02307|T02307 probable membrane transporter
At2g34190 [imported] - Arabidopsis thaliana, partial
(63%)
Length = 1165
Score = 33.5 bits (75), Expect = 0.20
Identities = 45/169 (26%), Positives = 71/169 (41%), Gaps = 15/169 (8%)
Frame = +3
Query: 291 LVTSLNWIAWFPYVLYDTDWMGLEVYGGKLGSKAYDAGVRAGALGLVLNSVV-------- 342
L++S WI + Y +W G+ +DAG G + VL S+V
Sbjct: 195 LISSAPWIK----IPYPLEW----------GAPTFDAGHSFGMMAAVLVSLVESTGAFKA 332
Query: 343 LGLMSLAVEPLGRYLG---GVKRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGGATIG- 398
++ A P L G + + ++N + T+ + V + G +G
Sbjct: 333 ASRLASATPPPAHVLSRGIGWQGIGILLNGLFGTLTGSTVSVENVG-----LLGSNRVGS 497
Query: 399 RPTPGVKAGALMFFAVLGIPLAITYSVPFAL-ASIYSSSTG--AGQGLS 444
R V AG ++FFA+LG A+ S+PF + A+IY G A GLS
Sbjct: 498 RRVIQVSAGFMIFFAMLGKFGALFASIPFPIFAAIYCVLFGLVASVGLS 644
>AL374845 similar to GP|12247468|gb| PnFL-1 {Ipomoea nil}, partial (25%)
Length = 460
Score = 32.7 bits (73), Expect = 0.34
Identities = 13/44 (29%), Positives = 26/44 (58%)
Frame = +1
Query: 2 APPPPPPNSTTQNNNSIAPSSFQVEPAQHAAGPSPLAKMIAVAS 45
+PPP P NS++ ++N+ S++ H++ P PL+ + + S
Sbjct: 94 SPPPTPQNSSSASSNT*PISAYSSPSLSHSSSPHPLSLLSVLTS 225
>TC89164 weakly similar to GP|10177340|dbj|BAB10596. transporter-like
protein {Arabidopsis thaliana}, partial (49%)
Length = 1129
Score = 32.3 bits (72), Expect = 0.45
Identities = 32/108 (29%), Positives = 46/108 (41%)
Frame = +3
Query: 101 TSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWILDV 160
+ R+GR+ P I G IAV I L G + GY M +I ++L
Sbjct: 552 SDRYGRK-PVIIMGIIAVVIFNTLFGLST--GYWMA---------------IITRFLLGS 677
Query: 161 ANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSY 208
N +L GP +A+ +L H I + S GVG ++G G Y
Sbjct: 678 LNGVL-GPVKAYASELFREEHQAIGLSTV--SAAWGVGLIIGPAIGGY 812
>TC86275 similar to PIR|E96603|E96603 unknown protein F14G9.26 [imported] -
Arabidopsis thaliana, partial (51%)
Length = 961
Score = 32.0 bits (71), Expect = 0.59
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = -2
Query: 3 PPPPPPNSTTQNNNSIAPSS 22
PPPPPP+S+++ N + PSS
Sbjct: 567 PPPPPPSSSSRGGNPLPPSS 508
>TC93020 similar to GP|6635840|gb|AAF20003.1| 1-acyl-sn-glycerol-3-phosphate
acyltransferase {Prunus dulcis}, partial (10%)
Length = 784
Score = 30.8 bits (68), Expect = 1.3
Identities = 16/34 (47%), Positives = 18/34 (52%)
Frame = +1
Query: 217 FTETKACDVFCANLKSCFFFSITLLLVLSGFALF 250
FT+TK D FC C FF TL +LS F F
Sbjct: 619 FTQTKKTDKFCYF**KCVFFFDTLKYLLSNFTYF 720
>AW736532 similar to GP|15983497|gb| AT5g38560/MBB18_10 {Arabidopsis
thaliana}, partial (9%)
Length = 626
Score = 30.4 bits (67), Expect = 1.7
Identities = 22/65 (33%), Positives = 30/65 (45%), Gaps = 6/65 (9%)
Frame = +3
Query: 3 PPPPPPNSTTQNNNSIAPSS-FQVEP-----AQHAAGPSPLAKMIAVASIAAGVQFGWAL 56
PPPPP NSTT + P+S Q+ P + SP + I+ +I A FG L
Sbjct: 78 PPPPPSNSTTPRLSPPTPASIIQLSPPPPPLQNGSENSSPQSGGISSGAILAIAAFGGIL 257
Query: 57 QLSLL 61
L +
Sbjct: 258 FLGFI 272
>TC79129 similar to PIR|A96725|A96725 hypothetical protein F20P5.7
[imported] - Arabidopsis thaliana, partial (64%)
Length = 1328
Score = 30.0 bits (66), Expect = 2.2
Identities = 11/21 (52%), Positives = 14/21 (66%)
Frame = +3
Query: 153 IGFWILDVANNMLQGPCRAFL 173
IGFWILD+ N + G C +L
Sbjct: 450 IGFWILDLYRNQMDGHCNFYL 512
>TC81547 similar to GP|13877739|gb|AAK43947.1 unknown protein {Arabidopsis
thaliana}, partial (4%)
Length = 725
Score = 29.6 bits (65), Expect = 2.9
Identities = 14/36 (38%), Positives = 22/36 (60%)
Frame = +3
Query: 3 PPPPPPNSTTQNNNSIAPSSFQVEPAQHAAGPSPLA 38
PPPPPP T+ ++SIA + + A + + P PL+
Sbjct: 27 PPPPPPAPTSDQSHSIANAPW----ASNHSAPPPLS 122
>AJ503819
Length = 294
Score = 29.6 bits (65), Expect = 2.9
Identities = 12/29 (41%), Positives = 19/29 (65%)
Frame = -3
Query: 226 FCANLKSCFFFSITLLLVLSGFALFYVHD 254
FC + + FFF L+L++S FA F+ H+
Sbjct: 289 FCWSFTTIFFFLFLLILLISIFASFFFHN 203
>BI264438 similar to GP|21427467|gb actin-related protein 6 {Arabidopsis
thaliana}, partial (43%)
Length = 695
Score = 29.6 bits (65), Expect = 2.9
Identities = 12/19 (63%), Positives = 13/19 (68%)
Frame = +1
Query: 3 PPPPPPNSTTQNNNSIAPS 21
PPPPPPN T+ S APS
Sbjct: 40 PPPPPPNKTSPPPPSAAPS 96
>TC91488 similar to GP|11078533|gb|AAG29095.1 Glu-tRNA(Gln) amidotransferase
subunit A {Arabidopsis thaliana}, partial (26%)
Length = 599
Score = 29.3 bits (64), Expect = 3.8
Identities = 16/52 (30%), Positives = 25/52 (47%), Gaps = 1/52 (1%)
Frame = +2
Query: 3 PPPPPPNSTTQNNNSIAPSSFQVEPAQH-AAGPSPLAKMIAVASIAAGVQFG 53
P PPP +S + S++P+ V + PS + M + IAAG + G
Sbjct: 95 PSPPPTSSNPTSPASVSPNPTSVPSSTSLPTNPSSIRHMTSTGRIAAGEEVG 250
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,054,234
Number of Sequences: 36976
Number of extensions: 268514
Number of successful extensions: 4671
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 2948
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4391
length of query: 515
length of database: 9,014,727
effective HSP length: 100
effective length of query: 415
effective length of database: 5,317,127
effective search space: 2206607705
effective search space used: 2206607705
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0134.21


