

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0134.16
(212 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
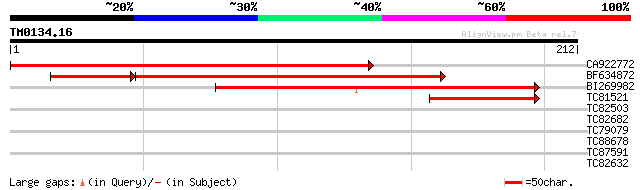
Score E
Sequences producing significant alignments: (bits) Value
CA922772 weakly similar to GP|7211993|gb|A ESTs gb|T76367 and gb... 181 2e-46
BF634872 139 1e-33
BI269982 weakly similar to GP|21593323|gb unknown {Arabidopsis t... 110 3e-25
TC81521 57 5e-09
TC82503 weakly similar to GP|18252325|gb|AAL66194.1 cytochrome P... 29 1.5
TC82682 similar to GP|20466394|gb|AAM20514.1 unknown protein {Ar... 27 4.4
TC79079 similar to GP|19698893|gb|AAL91182.1 splicing factor pu... 27 5.8
TC88678 similar to PIR|S51171|S51171 amino acid transport protei... 27 5.8
TC87591 similar to GP|9759578|dbj|BAB11141.1 3-hydroxyisobutyryl... 27 7.5
TC82632 homologue to GP|18086370|gb|AAL57645.1 At2g47390/T8I13.2... 26 9.8
>CA922772 weakly similar to GP|7211993|gb|A ESTs gb|T76367 and gb|AA404955
come from this gene. {Arabidopsis thaliana}, partial
(16%)
Length = 804
Score = 181 bits (459), Expect = 2e-46
Identities = 91/136 (66%), Positives = 113/136 (82%)
Frame = +2
Query: 1 VKGMQVGLTVNLKNQEGTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASSV 60
V+ +QVGLTVNL+NQEGTLKL LLDYGC VG+LSIK++GG AWLYQ+LVDAF GNI S+V
Sbjct: 374 VEDLQVGLTVNLRNQEGTLKLILLDYGCDVGELSIKMNGGAAWLYQVLVDAFKGNIGSAV 553
Query: 61 EEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFTER 120
E+AVS+KI EGI LD LQ+LPK I +D+T+ALN+SFV NPVLSNSSI + I+GLFTER
Sbjct: 554 EDAVSKKIREGIPTLDNLLQTLPKTISIDETAALNISFVDNPVLSNSSIELEIDGLFTER 733
Query: 121 SEVVESEGYKKGFKIS 136
++V+ + Y + IS
Sbjct: 734 NDVLVPQVYHRRSDIS 781
>BF634872
Length = 688
Score = 139 bits (349), Expect = 1e-33
Identities = 72/116 (62%), Positives = 90/116 (77%)
Frame = +1
Query: 48 LVDAFGGNIASSVEEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNS 107
LVDAF GNI S+VE+AVS+KI EGI LD LQ+LPK I +D+T+ALN+SFV NPVLSNS
Sbjct: 232 LVDAFKGNIGSAVEDAVSKKIREGIPTLDNLLQTLPKTISIDETAALNISFVDNPVLSNS 411
Query: 108 SIAIAINGLFTERSEVVESEGYKKGFKISSACGGLPNMIKVSLHEYVIQSASLVYF 163
SI + I+GLFTER++V+ + Y + IS + GGLP MI +SLHE V +SAS VYF
Sbjct: 412 SIELEIDGLFTERNDVLVPQVYHRRSDISVSSGGLPKMINISLHENVFKSASEVYF 579
Score = 57.0 bits (136), Expect = 5e-09
Identities = 25/32 (78%), Positives = 29/32 (90%)
Frame = +2
Query: 16 EGTLKLSLLDYGCYVGDLSIKLDGGTAWLYQL 47
EGTLKL LLDYGC VG+LSIK++GG AWLYQ+
Sbjct: 2 EGTLKLILLDYGCDVGELSIKMNGGAAWLYQV 97
>BI269982 weakly similar to GP|21593323|gb unknown {Arabidopsis thaliana},
partial (30%)
Length = 651
Score = 110 bits (276), Expect = 3e-25
Identities = 56/122 (45%), Positives = 83/122 (67%), Gaps = 1/122 (0%)
Frame = +2
Query: 78 FLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFTERSEVVESEG-YKKGFKIS 136
+L+SLPK++P+D S+LNV+FV N +LS+SSI NGLF +R++ + + K K+
Sbjct: 2 YLKSLPKEVPVDDHSSLNVTFVNNVLLSDSSIGFETNGLFIKRNDSLPIPNLWHKNSKLP 181
Query: 137 SACGGLPNMIKVSLHEYVIQSASLVYFNAGKMQLIIDELPDQVILNTAECRFIVPQLYKQ 196
C M+ +SL E V SAS +Y++A M I+D++PDQ +LNTA RFI+PQLY++
Sbjct: 182 ILCTNSSKMLAISLDEAVFNSASSLYYDAKFMHWIVDKIPDQSLLNTAGWRFIIPQLYRK 361
Query: 197 YP 198
YP
Sbjct: 362 YP 367
>TC81521
Length = 869
Score = 57.0 bits (136), Expect = 5e-09
Identities = 25/41 (60%), Positives = 33/41 (79%)
Frame = +1
Query: 158 ASLVYFNAGKMQLIIDELPDQVILNTAECRFIVPQLYKQYP 198
AS VYF A +Q I+DELP+Q +LNTA+ + ++PQLYKQYP
Sbjct: 1 ASEVYFAADALQWILDELPNQALLNTADWKILIPQLYKQYP 123
>TC82503 weakly similar to GP|18252325|gb|AAL66194.1 cytochrome P450 {Pyrus
communis}, partial (39%)
Length = 806
Score = 28.9 bits (63), Expect = 1.5
Identities = 28/128 (21%), Positives = 58/128 (44%), Gaps = 4/128 (3%)
Frame = -2
Query: 82 LPKQIP-LDKTSALNVSFVGNPVLSNSSIAIAINGLFTERSEVVESEGYKKGFKISSACG 140
LP P LD + ++ F + ++ + S+A+ F + S+ S+G K +S
Sbjct: 532 LPPNAP*LDLIAIQDIPFFQSQMV*DESVALQHLCCFLQISQAKTSQGQSKNLDLS*LPM 353
Query: 141 GLPNMIKVSLHEYVIQSASLVYFNAGKMQLIIDELPDQVI---LNTAECRFIVPQLYKQY 197
P + + + ++Q +L + + + +DE ++ NT ++P L+ Y
Sbjct: 352 HSP*SLSF*V*K-ILQQCNLPWIGVVLIGVPLDET*GFLL*PCSNTPTSPNLIPPLFSSY 176
Query: 198 PYVFDTMF 205
P +F T+F
Sbjct: 175 PPLFPTLF 152
>TC82682 similar to GP|20466394|gb|AAM20514.1 unknown protein {Arabidopsis
thaliana}, partial (31%)
Length = 1243
Score = 27.3 bits (59), Expect = 4.4
Identities = 10/29 (34%), Positives = 16/29 (54%)
Frame = -1
Query: 28 CYVGDLSIKLDGGTAWLYQLLVDAFGGNI 56
C+ S+ ++GGT+WL LL F +
Sbjct: 646 CFTDMFSLAIEGGTSWLLPLLSVVFSSQL 560
>TC79079 similar to GP|19698893|gb|AAL91182.1 splicing factor putative
{Arabidopsis thaliana}, partial (17%)
Length = 1114
Score = 26.9 bits (58), Expect = 5.8
Identities = 31/129 (24%), Positives = 56/129 (43%), Gaps = 17/129 (13%)
Frame = +1
Query: 54 GNIASSVEEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIA- 112
GN+ V E + ++E + L + + Q+P +K G P ++++A
Sbjct: 511 GNLKGQVLEITVQSLSETVGSLKEKIAG-EIQLPANKQK-----LSGKPGFLKDNMSLAH 672
Query: 113 --INGLFTERSEVVESEG---------YKKGFKISSACGGLPNMI-----KVSLHEYVIQ 156
++G T + E G +KK F IS++ GL +I LHE+ ++
Sbjct: 673 YNVSGGETLSLALRERGGRKR*AYYALFKKVFVISASL*GLSILILDGLHNAYLHEFYVE 852
Query: 157 SASLVYFNA 165
L+YF+A
Sbjct: 853 YLYLLYFDA 879
>TC88678 similar to PIR|S51171|S51171 amino acid transport protein AAT1 -
Arabidopsis thaliana, partial (72%)
Length = 1520
Score = 26.9 bits (58), Expect = 5.8
Identities = 19/47 (40%), Positives = 27/47 (57%), Gaps = 2/47 (4%)
Frame = -2
Query: 63 AVSEKIN--EGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNS 107
A SEKIN E +LD+ L+ + K S V+F+G PVLS++
Sbjct: 1345 ATSEKINKVEIDKRLDRILRLVKKATIAVAISIEIVAFIGVPVLSST 1205
>TC87591 similar to GP|9759578|dbj|BAB11141.1 3-hydroxyisobutyryl-coenzyme A
hydrolase {Arabidopsis thaliana}, partial (86%)
Length = 1470
Score = 26.6 bits (57), Expect = 7.5
Identities = 23/101 (22%), Positives = 39/101 (37%)
Frame = +3
Query: 26 YGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASSVEEAVSEKINEGIAKLDKFLQSLPKQ 85
+G YVG +LDG L + SS+EE++ + A + + +Q
Sbjct: 534 FGEYVGLTGARLDGAEMLACGLATHFVPSSKLSSLEESLCKVETSDPAIVSAIIDKYSEQ 713
Query: 86 IPLDKTSALNVSFVGNPVLSNSSIAIAINGLFTERSEVVES 126
L K S + + N S ++ + L TE +S
Sbjct: 714 PSLKKDSVYHRMDIINKCFSRKTVEDILYSLETEAMSKADS 836
>TC82632 homologue to GP|18086370|gb|AAL57645.1 At2g47390/T8I13.23
{Arabidopsis thaliana}, partial (26%)
Length = 848
Score = 26.2 bits (56), Expect = 9.8
Identities = 11/34 (32%), Positives = 19/34 (55%)
Frame = -2
Query: 176 PDQVILNTAECRFIVPQLYKQYPYVFDTMFLIML 209
P + + N ++C VP+L P++ FLI+L
Sbjct: 643 PFRALKNLSDCMVNVPELLSSSPWISKIGFLILL 542
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,590,441
Number of Sequences: 36976
Number of extensions: 64390
Number of successful extensions: 306
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 304
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 305
length of query: 212
length of database: 9,014,727
effective HSP length: 92
effective length of query: 120
effective length of database: 5,612,935
effective search space: 673552200
effective search space used: 673552200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0134.16


