

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
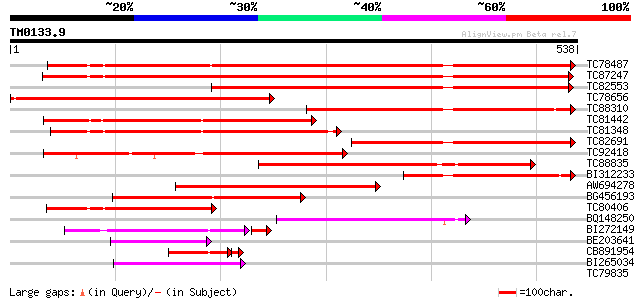
Query= TM0133.9
(538 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78487 similar to PIR|A84663|A84663 probable beta-ketoacyl-CoA ... 614 e-176
TC87247 similar to GP|9714501|emb|CAC01441.1 putative fatty acid... 577 e-165
TC82553 similar to PIR|T00951|T00951 probable 3-oxoacyl-[acyl-ca... 476 e-134
TC78656 similar to GP|18447765|gb|AAL67993.1 fiddlehead-like pro... 448 e-126
TC88310 similar to PIR|F86327|F86327 protein F18O14.21 [imported... 343 1e-94
TC81442 similar to PIR|F86327|F86327 protein F18O14.21 [imported... 284 7e-77
TC81348 similar to GP|17979464|gb|AAL50069.1 At1g68530/T26J14_10... 261 4e-70
TC82691 similar to PIR|A84663|A84663 probable beta-ketoacyl-CoA ... 261 4e-70
TC92418 similar to GP|3551960|gb|AAC34858.1| senescence-associat... 248 3e-66
TC88835 similar to GP|16226847|gb|AAL16279.1 At1g07720/F24B9_16 ... 221 4e-58
BI312233 homologue to GP|17979464|gb At1g68530/T26J14_10 {Arabid... 213 2e-55
AW694278 similar to PIR|T48449|T48 fatty acid elongase-like prot... 172 2e-43
BG456193 similar to PIR|A84663|A846 probable beta-ketoacyl-CoA s... 166 2e-41
TC80406 weakly similar to GP|9714501|emb|CAC01441.1 putative fat... 142 2e-34
BQ148250 similar to GP|21536949|gb putative fatty acid elongase ... 136 2e-32
BI272149 weakly similar to GP|9714501|emb| putative fatty acid e... 111 4e-29
BE203641 weakly similar to PIR|F86141|F861 protein T25K16.11 [im... 69 3e-12
CB891954 similar to PIR|A84663|A846 probable beta-ketoacyl-CoA s... 62 8e-11
BI265034 similar to GP|21536949|gb putative fatty acid elongase ... 53 3e-07
TC79835 homologue to SP|P51082|CHSB_PEA Chalcone synthase 1B (EC... 38 0.009
>TC78487 similar to PIR|A84663|A84663 probable beta-ketoacyl-CoA synthase
[imported] - Arabidopsis thaliana, partial (97%)
Length = 1999
Score = 614 bits (1584), Expect = e-176
Identities = 295/501 (58%), Positives = 388/501 (76%)
Frame = +1
Query: 37 RRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDAS 96
R LPDF +SV LKYVKLGYHYLI HG+YLF PL++++ +A++ + S D++ +WE+
Sbjct: 280 RILPDFKKSVKLKYVKLGYHYLITHGMYLFLSPLVVLI-AAQLSTFSLNDIYD-IWENLQ 453
Query: 97 YDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKFD 156
Y+L SV+ + VF + YFM++PRP+YL++F+CY+PE+ K ++ +++ + SG F
Sbjct: 454 YNLVSVIICSTLLVFLSTFYFMTRPRPVYLVNFSCYKPEESRKCTKRIFMDHSRASGFFT 633
Query: 157 EESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGI 216
EE+L FQ++IL SG+G+ TY+P+AV+S N + MKE R EA VMFGA+DELF KT +
Sbjct: 634 EENLDFQRKILERSGLGENTYLPEAVLSIPPNPS-MKEARKEAEAVMFGAIDELFSKTTV 810
Query: 217 RPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQ 276
+PKDIGIL+VNCS+F PTPSLSAMIINHYK+RGNI+SYNLGGMGCSAGII +DLA+++LQ
Sbjct: 811 KPKDIGILIVNCSLFCPTPSLSAMIINHYKLRGNIMSYNLGGMGCSAGIISIDLAKELLQ 990
Query: 277 SNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHI 336
+PN+YA+VVS E + NWY G +RS L+ NC FRMG +A+LLSNR D R+KY+L H
Sbjct: 991 VHPNSYALVVSMENITLNWYPGNDRSKLVSNCLFRMGGAAILLSNRSSDRRRSKYQLVHT 1170
Query: 337 VRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLL 396
VRT+KGADD+ F CV QEEDD G+ +SKDL+ + GDALKTNITTLGPLVLP SEQLL
Sbjct: 1171VRTNKGADDKCFSCVTQEEDDNGKVGVTLSKDLMAVAGDALKTNITTLGPLVLPTSEQLL 1350
Query: 397 FFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEK 456
FFATLV + LF K KPYIP++KLAFEHFC+HA + +LDEL++NL+LS
Sbjct: 1351FFATLVGKKLFKMKI---------KPYIPDFKLAFEHFCIHAGGRAVLDELEKNLKLSPW 1503
Query: 457 NMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKR 516
+ME SRMTL+RFGNTSSSS+WYELAY EA R+++GDR WQ+AFGSGFKCNS VW +++
Sbjct: 1504HMEPSRMTLYRFGNTSSSSLWYELAYTEANGRIKKGDRTWQIAFGSGFKCNSAVWKALRT 1683
Query: 517 VNKPSRNNPWLDCINRYPVSL 537
+N NPW+D I+++PV +
Sbjct: 1684INPAKEKNPWIDEIHQFPVDV 1746
>TC87247 similar to GP|9714501|emb|CAC01441.1 putative fatty acid elongase
{Zea mays}, partial (91%)
Length = 2102
Score = 577 bits (1486), Expect = e-165
Identities = 284/504 (56%), Positives = 373/504 (73%)
Frame = +3
Query: 32 SVRVRRRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKL 91
S + RR+LP+FL SV LKYVKLGYHYLI+ +YL IPL SA + ++S + + L
Sbjct: 210 SPQERRKLPNFLLSVRLKYVKLGYHYLISKAMYLLLIPLFGAA-SAHLSTISYHVIIQ-L 383
Query: 92 WEDASYDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASK 151
+E+ ++L SV ++ VF +++YFMS+PR +YL+DFACY+P+ E +RE ++ +
Sbjct: 384 YENLKFNLVSVTLCTSLMVFLVTLYFMSRPRGVYLVDFACYKPKQEYTCTREIFMNRSEL 563
Query: 152 SGKFDEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELF 211
+G F +E+LAFQK+IL + + + +T + + FGA+DEL
Sbjct: 564 TGTFSDENLAFQKKILEKVWFRSKDLFAPSNIECSTKSLYG*SKERSWRKLWFGAIDELL 743
Query: 212 EKTGIRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLA 271
EKTG++ KDIGILVVNCS+FNPTPSLSAMI+NHYK+RGN+ SYNLGGMGCSAG+I +DLA
Sbjct: 744 EKTGVKAKDIGILVVNCSLFNPTPSLSAMIVNHYKLRGNVQSYNLGGMGCSAGLISIDLA 923
Query: 272 RDILQSNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKY 331
+ +LQ +PN+YA+VVS E + NWY G +RSML+ NC FRMG SA+LLSN+ D RAKY
Sbjct: 924 KQLLQVHPNSYALVVSMENITLNWYFGNDRSMLVSNCLFRMGGSAILLSNKASDSKRAKY 1103
Query: 332 RLEHIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPF 391
+L H VRTHKGADDRS+ CV+QEED++K G+ +SKDL+ + G+ALKTNITTLGPLVLP
Sbjct: 1104QLIHTVRTHKGADDRSYGCVFQEEDEKKSIGVSLSKDLMAVAGEALKTNITTLGPLVLPM 1283
Query: 392 SEQLLFFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNL 451
SEQLLFFATLV R F K KPYIP++KLAFEHFC+HA + +LDEL++NL
Sbjct: 1284SEQLLFFATLVARKAFKMKI---------KPYIPDFKLAFEHFCIHAGGRAVLDELEKNL 1436
Query: 452 ELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVW 511
+LS+ +ME SRMTL+RFGNTSSSS+WYELAY EAK R+R+GDR WQ+AFGSGFKCNS VW
Sbjct: 1437DLSDWHMEPSRMTLNRFGNTSSSSLWYELAYTEAKGRIRKGDRTWQIAFGSGFKCNSAVW 1616
Query: 512 LSMKRVNKPSRNNPWLDCINRYPV 535
+++ V+ NPW+D I+ +PV
Sbjct: 1617RALRTVDPAKEKNPWMDEIHEFPV 1688
>TC82553 similar to PIR|T00951|T00951 probable
3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41)
F20D22.1 - Arabidopsis thaliana, partial (65%)
Length = 1343
Score = 476 bits (1224), Expect = e-134
Identities = 224/344 (65%), Positives = 279/344 (80%)
Frame = +3
Query: 192 MKEGRAEASTVMFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNI 251
M E R EA VMFGA+DE+ +KTG++ KDIGILVVNCS+FNPTPSLSAMI+NHYK+RGNI
Sbjct: 27 MAEARKEAEEVMFGAIDEVLQKTGVKAKDIGILVVNCSLFNPTPSLSAMIVNHYKLRGNI 206
Query: 252 LSYNLGGMGCSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFR 311
LSYNLGGMGCSAG+I +DLA+ +LQ +PN+YA+VVS E + NWY G +RSML+PNC FR
Sbjct: 207 LSYNLGGMGCSAGLISIDLAKQLLQVHPNSYALVVSMENITLNWYFGNDRSMLVPNCLFR 386
Query: 312 MGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIE 371
MG +A+LLSN+ D R+KY+L H VRTHKGAD++S+ CV+QEEDD K G+ +SKDL+
Sbjct: 387 MGGAAVLLSNKPRDRLRSKYQLVHTVRTHKGADNKSYGCVFQEEDDTKQVGVSLSKDLMA 566
Query: 372 IGGDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAF 431
+ G+ALKTNITTLGPLVLP SEQLLFF TLV R +F K KPYIP++KLAF
Sbjct: 567 VAGEALKTNITTLGPLVLPMSEQLLFFGTLVARKIFKMKI---------KPYIPDFKLAF 719
Query: 432 EHFCVHAASKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRR 491
EHFC+HA + +LDEL++NL+LS+ +ME SRMTL+RFGNTSSSS+WYELAY EAK R+++
Sbjct: 720 EHFCIHAGGRAVLDELEKNLDLSDWHMEPSRMTLNRFGNTSSSSLWYELAYTEAKGRIKK 899
Query: 492 GDRVWQLAFGSGFKCNSVVWLSMKRVNKPSRNNPWLDCINRYPV 535
GDR WQ+AFGSGFKCNS VW ++K +N NPW+D I+ +PV
Sbjct: 900 GDRTWQIAFGSGFKCNSAVWKALKTINPAKEKNPWIDEIHEFPV 1031
>TC78656 similar to GP|18447765|gb|AAL67993.1 fiddlehead-like protein
{Gossypium hirsutum}, partial (46%)
Length = 806
Score = 448 bits (1152), Expect = e-126
Identities = 223/251 (88%), Positives = 240/251 (94%)
Frame = +2
Query: 1 MARNSERDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 60
MA N E DMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN
Sbjct: 56 MATN-EGDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 232
Query: 61 HGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLASVLASFAVFVFTLSVYFMSK 120
HG+YLFTIP+LLVVFSAEVGSLSKEDLWKK+WEDA+YDLASVL+S AVFVFT ++YFMS+
Sbjct: 233 HGVYLFTIPILLVVFSAEVGSLSKEDLWKKIWEDATYDLASVLSSLAVFVFTFTLYFMSR 412
Query: 121 PRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEESLAFQKRILMSSGIGDETYIPK 180
PRPIYLIDFACY+P+DELKVSREQ IELA KSGKFDE SL FQKRI+MSSGIGDETYIP+
Sbjct: 413 PRPIYLIDFACYQPDDELKVSREQLIELARKSGKFDEGSLEFQKRIVMSSGIGDETYIPR 592
Query: 181 AVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAM 240
+V+SS+ NTATMKEGRAEAS VMFGALDELFEKTGIRPKD+G+LVVNCSIFNPTPSLSAM
Sbjct: 593 SVISSSENTATMKEGRAEASMVMFGALDELFEKTGIRPKDVGVLVVNCSIFNPTPSLSAM 772
Query: 241 IINHYKMRGNI 251
IINHYKMRGNI
Sbjct: 773 IINHYKMRGNI 805
>TC88310 similar to PIR|F86327|F86327 protein F18O14.21 [imported] -
Arabidopsis thaliana, partial (47%)
Length = 995
Score = 343 bits (879), Expect = 1e-94
Identities = 165/256 (64%), Positives = 205/256 (79%)
Frame = +3
Query: 282 YAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHK 341
YAVVVSTE + NWY G ++SMLIPNC FR+G +A+LLSN+ D +RAKY+L H+VRTHK
Sbjct: 3 YAVVVSTENITQNWYFGNKKSMLIPNCLFRVGGAAVLLSNKGCDRSRAKYKLVHVVRTHK 182
Query: 342 GADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATL 401
GADD++F+CVYQE+DD G+ +SKDL+ I G ALKTNITTLGPLVLP SEQLLFF TL
Sbjct: 183 GADDKAFKCVYQEQDDVGKTGVSLSKDLMAIAGGALKTNITTLGPLVLPVSEQLLFFTTL 362
Query: 402 VWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEAS 461
V + F K+ KPYIP++KLAFEHFC+HA + ++DEL++NL+L ++EAS
Sbjct: 363 VIKKWFNAKT---------KPYIPDFKLAFEHFCIHAGGRAVIDELEKNLQLMPDHVEAS 515
Query: 462 RMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKRVNKPS 521
RMTLHRFGNTSSSSIWYELAY+EAK R+R+G+R+WQ+AFGSGFKCNS VW +MK V K S
Sbjct: 516 RMTLHRFGNTSSSSIWYELAYIEAKGRMRKGNRIWQIAFGSGFKCNSAVWQAMKHV-KAS 692
Query: 522 RNNPWLDCINRYPVSL 537
+PW DCI+RYPV +
Sbjct: 693 PMSPWEDCIDRYPVEI 740
>TC81442 similar to PIR|F86327|F86327 protein F18O14.21 [imported] -
Arabidopsis thaliana, partial (47%)
Length = 915
Score = 284 bits (726), Expect = 7e-77
Identities = 143/259 (55%), Positives = 186/259 (71%)
Frame = +2
Query: 33 VRVRRRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLW 92
+ RRLPDFLQSVNLKYVKLGYHYLI H + L IPL+ V+ +V ++ ++ +LW
Sbjct: 140 IHTNRRLPDFLQSVNLKYVKLGYHYLITHLLTLCLIPLMSVII-IQVSQMNPNEI-HQLW 313
Query: 93 EDASYDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKS 152
Y+L S++ A+ VF +VY M++PR IYLIDF+CY+P L V ++I+ +
Sbjct: 314 LHLKYNLVSIITCSAILVFGSTVYIMTRPRSIYLIDFSCYKPPSNLAVKFTKFIQHSKLK 493
Query: 153 GKFDEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFE 212
G FDE SL FQ++IL SG+G++TY+P+A+ T M R EA VM+GALD LF
Sbjct: 494 GDFDESSLEFQRKILERSGLGEDTYLPEAM-HKIPPTPCMASAREEAEQVMYGALDNLFA 670
Query: 213 KTGIRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLAR 272
T I+PKDIGILVVNCS+FNPTPSLSAMI+N YK+RGNI S+NLGGMGCSAG+I +DLA+
Sbjct: 671 NTKIKPKDIGILVVNCSLFNPTPSLSAMIVNKYKLRGNIRSFNLGGMGCSAGVIAIDLAK 850
Query: 273 DILQSNPNNYAVVVSTEMV 291
D+LQ + N YAVV STE +
Sbjct: 851 DMLQVHRNTYAVVDSTENI 907
>TC81348 similar to GP|17979464|gb|AAL50069.1 At1g68530/T26J14_10
{Arabidopsis thaliana}, partial (53%)
Length = 906
Score = 261 bits (668), Expect = 4e-70
Identities = 139/277 (50%), Positives = 190/277 (68%)
Frame = +2
Query: 39 LPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYD 98
LPDF SV LKYVKLGY YL+NH I L IP+++ V S E+ L ++ LW + +
Sbjct: 92 LPDFSSSVKLKYVKLGYQYLVNHIITLTLIPIMIGV-SIELIRLGPNEILN-LWNSLNLN 265
Query: 99 LASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEE 158
L +L S + +F +VYFMSKPR IYL+D+AC++P +V ++E + K + +
Sbjct: 266 LVQILCSSFLVIFIATVYFMSKPRTIYLVDYACFKPPVTCRVPFATFMEHSRLILKNNPK 445
Query: 159 SLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRP 218
S+ FQ RIL SG+G+ET +P ++ TM+ R EA V+F A+D LFEKTG++P
Sbjct: 446 SVEFQMRILERSGLGEETCLPPSI-HYIPPKPTMESARGEAELVIFSAMDSLFEKTGLKP 622
Query: 219 KDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQSN 278
KDI IL+VNCS+F+PTPSLSAM+IN YK+R NI S+NL GMGCSAG+I +DLARD+LQ +
Sbjct: 623 KDIDILIVNCSLFSPTPSLSAMVINKYKLRSNIRSFNLSGMGCSAGLISIDLARDLLQVH 802
Query: 279 PNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCS 315
PN+ AVVVSTE++ N+YQGK +S + F + CS
Sbjct: 803 PNSNAVVVSTEIITPNYYQGK*KS----HAFTKFVCS 901
>TC82691 similar to PIR|A84663|A84663 probable beta-ketoacyl-CoA synthase
[imported] - Arabidopsis thaliana, partial (41%)
Length = 823
Score = 261 bits (668), Expect = 4e-70
Identities = 126/213 (59%), Positives = 160/213 (74%)
Frame = +2
Query: 325 DYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTL 384
D R+KY+L VRT+K +DD+ F CV QEED G+ +SKDL+ + GDALKTNITTL
Sbjct: 26 DRRRSKYQLITTVRTNKASDDKCFSCVTQEEDANGKIGVTLSKDLMAVAGDALKTNITTL 205
Query: 385 GPLVLPFSEQLLFFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPIL 444
GPLVLP SEQLLFF TLV + LF K KPYIP++KLAFEHFC+HA + +L
Sbjct: 206 GPLVLPTSEQLLFFTTLVGKKLFKMKI---------KPYIPDFKLAFEHFCIHAGGRAVL 358
Query: 445 DELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGF 504
DEL++NL+LS +ME SRMTL+RFGNTSSSS+WYELAY EAK R+++GDR WQ+AFGSGF
Sbjct: 359 DELEKNLQLSPWHMEPSRMTLYRFGNTSSSSLWYELAYTEAKGRIKKGDRTWQIAFGSGF 538
Query: 505 KCNSVVWLSMKRVNKPSRNNPWLDCINRYPVSL 537
KCNS VW +++ +N +PW+D I ++PV +
Sbjct: 539 KCNSAVWKALRTINPVKEKSPWIDEIGQFPVDV 637
>TC92418 similar to GP|3551960|gb|AAC34858.1| senescence-associated protein
15 {Hemerocallis hybrid cultivar}, partial (35%)
Length = 1034
Score = 248 bits (634), Expect = 3e-66
Identities = 141/301 (46%), Positives = 189/301 (61%), Gaps = 13/301 (4%)
Frame = +2
Query: 33 VRVRRRLPDFLQSVNLKYVKLGYHYLINHG---IYLFTIPLLL-VVFSAEVGSLSKEDLW 88
+++RRRLPDFLQ+V LKYVKLGY Y N I+ F +PLLL + ++ L L
Sbjct: 128 IKIRRRLPDFLQTVKLKYVKLGYGYSCNATTILIFTFVVPLLLFLTVQFQLTDLKLHRLS 307
Query: 89 KKLWEDA-SYDLASVLAS-FAVFVFTLSVYFMSKPRPIYLIDFACYRPED-------ELK 139
+ L E D + + S F +F+F L Y+ + PIYL+DFACY+PE+ +
Sbjct: 308 QLLLEHTVQLDTDTAIGSIFLLFLFGL--YYAKRSPPIYLVDFACYKPEN*AENCS*IFR 481
Query: 140 VSREQYIELASKSGKFDEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEA 199
+ + + ++ + ++M E MKE R EA
Sbjct: 482 *NVRR*WRIRRRNTSISTPHFVKSRSLVMKLPCQTE*-------CQVHQNLCMKEARLEA 640
Query: 200 STVMFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGM 259
+VMFG+LD LF KTG+ P+DI ILVVNCS+FNPTPSLSAMI+NHYK+R NI SYNLGGM
Sbjct: 641 ESVMFGSLDALFAKTGVNPRDIDILVVNCSLFNPTPSLSAMIVNHYKLRTNIKSYNLGGM 820
Query: 260 GCSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLL 319
GCSAG+I +DLA+D+L++NPN+YAVV+STE + NWY G +RSML+ NC FRMG +A+LL
Sbjct: 821 GCSAGLISIDLAKDLLKANPNSYAVVLSTENLTLNWYFGNDRSMLLSNCIFRMGGAAILL 1000
Query: 320 S 320
S
Sbjct: 1001S 1003
>TC88835 similar to GP|16226847|gb|AAL16279.1 At1g07720/F24B9_16
{Arabidopsis thaliana}, partial (51%)
Length = 788
Score = 221 bits (564), Expect = 4e-58
Identities = 112/263 (42%), Positives = 168/263 (63%)
Frame = +3
Query: 237 LSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWY 296
LS+ IIN YKMR ++ YNL MGCSA +I +D+ ++I +S N A++V++E + NWY
Sbjct: 3 LSSRIINRYKMRHDVKVYNLTAMGCSASLISLDIVKNIFKSQRNKLALLVTSESLCPNWY 182
Query: 297 QGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEED 356
G ++SM++ NC FR G A+LL+N+R NR+ +L+ +VRTH GA D S+ C Q+ED
Sbjct: 183 TGNDKSMILANCLFRSGGCAVLLTNKRSLKNRSILKLKCLVRTHHGARDDSYTCCTQKED 362
Query: 357 DQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFGGKSDGNSS 416
+Q G+ + K L + A N+ + P +LP E L F + + L K +
Sbjct: 363 EQGRLGIHLGKTLPKAATRAFVDNLRVISPKILPTKEILRFLFVTLLKKL---KKTSGGT 533
Query: 417 PSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSI 476
S K P N+K +HFC+H K ++D + +L+LSE ++E +RMTLHRFGNTS+SS+
Sbjct: 534 KSTKSPL--NFKTGVDHFCLHTGGKAVIDGIGMSLDLSEYDLEPARMTLHRFGNTSASSL 707
Query: 477 WYELAYMEAKERVRRGDRVWQLA 499
WY L YMEAK+R+++GDRV ++
Sbjct: 708 WYVLXYMEAKKRLKKGDRVXMIS 776
>BI312233 homologue to GP|17979464|gb At1g68530/T26J14_10 {Arabidopsis
thaliana}, partial (32%)
Length = 744
Score = 213 bits (541), Expect = 2e-55
Identities = 100/164 (60%), Positives = 127/164 (76%)
Frame = +1
Query: 374 GDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEH 433
G ALK+NITT+GPLVLP SEQLLF TL+ R +F K KPYIP++K AFEH
Sbjct: 1 GKALKSNITTMGPLVLPASEQLLFLITLIGRKIFNPKW---------KPYIPDFKQAFEH 153
Query: 434 FCVHAASKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGD 493
FC+HA + ++DELQ+NL+LS +++EASRMTLHRFGNTSSSS+WYEL Y+E+K R+++GD
Sbjct: 154 FCIHAGGRAVIDELQKNLQLSTEHVEASRMTLHRFGNTSSSSLWYELNYIESKGRMKKGD 333
Query: 494 RVWQLAFGSGFKCNSVVWLSMKRVNKPSRNNPWLDCINRYPVSL 537
RVWQ+AFGSGFKCNS VW + + P + PW DCI+RYPV +
Sbjct: 334 RVWQIAFGSGFKCNSAVWKCNRSIKTPI-DGPWTDCIDRYPVHI 462
>AW694278 similar to PIR|T48449|T48 fatty acid elongase-like protein -
Arabidopsis thaliana, partial (39%)
Length = 641
Score = 172 bits (437), Expect = 2e-43
Identities = 81/195 (41%), Positives = 126/195 (64%)
Frame = +3
Query: 158 ESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIR 217
E F + ++SSGIG+ TY P+ V+ T+K+ E +MF LD LF+KT
Sbjct: 51 EEFRFLLKTMVSSGIGENTYCPRNVLEGRETCPTLKDTYEEIDEIMFDTLDNLFKKTCFS 230
Query: 218 PKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQS 277
P +I ILVVN S+F+P PSL+A IIN YKMR ++ +NL GMGCSA ++ +DL + + ++
Sbjct: 231 PSEIDILVVNVSLFSPAPSLTARIINRYKMREDVKVFNLAGMGCSASVVAIDLVQQLFKT 410
Query: 278 NPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIV 337
N+ +VVSTE +G +WY GK++ M++ NC FR G ++L +N+ +RA +L+H+
Sbjct: 411 YENSLGIVVSTEDLGSHWYCGKDKKMMLSNCLFRAGGCSMLFTNKTELKDRAILKLKHME 590
Query: 338 RTHKGADDRSFRCVY 352
RT G+DD + + ++
Sbjct: 591 RTQYGSDDEALQLLH 635
>BG456193 similar to PIR|A84663|A846 probable beta-ketoacyl-CoA synthase
[imported] - Arabidopsis thaliana, partial (16%)
Length = 643
Score = 166 bits (420), Expect = 2e-41
Identities = 80/183 (43%), Positives = 123/183 (66%)
Frame = +1
Query: 98 DLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDE 157
++ S+L A + ++ Y + +YL+DFACY+P S+E +I+ G F +
Sbjct: 97 NIFSLLMWCASISYIVTFYQKKCSKKVYLVDFACYKPFPNGICSKELFIKQTKSGGNFKD 276
Query: 158 ESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIR 217
ES+ FQK+IL SG GD+TY+P++++ N + + E R E +V+FGA+++L KT ++
Sbjct: 277 ESIDFQKKILDRSGFGDKTYVPESLLKIPQNISIV-EARKETESVIFGAINDLLLKTKMK 453
Query: 218 PKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQS 277
+DI IL+ NCSIFNP PSLSAM++NH+K++ IL YNL GMGCSA +I +DLA+ +LQ
Sbjct: 454 AEDIEILITNCSIFNPVPSLSAMVVNHFKLKHTILCYNLSGMGCSAXLIAIDLAKQLLQV 633
Query: 278 NPN 280
+ N
Sbjct: 634 HQN 642
>TC80406 weakly similar to GP|9714501|emb|CAC01441.1 putative fatty acid
elongase {Zea mays}, partial (30%)
Length = 726
Score = 142 bits (359), Expect = 2e-34
Identities = 72/161 (44%), Positives = 113/161 (69%)
Frame = +3
Query: 36 RRRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDA 95
R +LP+FL SV LKYVKLGYHYLI++ +YL IPLL V S + + S +DL + ++E+
Sbjct: 228 RSKLPNFLISVKLKYVKLGYHYLISNAMYLVLIPLLGVA-SVHLSTFSIKDLIQ-IYENL 401
Query: 96 SYDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKF 155
++ S+L ++ VF ++YFMS+PR IYL+DFACY+P+ +L+V+RE ++E + + F
Sbjct: 402 KFNFVSMLICTSLVVFLATLYFMSRPRGIYLVDFACYKPQKDLQVTREIFVERSILTKAF 581
Query: 156 DEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGR 196
EE+L+FQK+IL SG+G +TY+P ++ +K+G+
Sbjct: 582 TEETLSFQKKILERSGLGQKTYLPPXIMRVPLIHVWLKQGK 704
>BQ148250 similar to GP|21536949|gb putative fatty acid elongase {Arabidopsis
thaliana}, partial (36%)
Length = 647
Score = 136 bits (342), Expect = 2e-32
Identities = 71/189 (37%), Positives = 109/189 (57%), Gaps = 5/189 (2%)
Frame = +1
Query: 254 YNLGGMGCSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMG 313
YNL GMGCSA +I +D+ ++I +S N A++V++E + NWY G +RSM++ NC FR G
Sbjct: 76 YNLTGMGCSASLISLDIVKNIFKSQRNKLALLVTSESLSPNWYTGNDRSMILANCLFRSG 255
Query: 314 CSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIG 373
++LL+N+R NR+ +L+ +VRTH GA + S+ C Q+ED+Q G + K L +
Sbjct: 256 GCSILLTNKRSLKNRSILKLKCLVRTHHGAREDSYNCCIQKEDEQGRLGFHLGKTLPKAA 435
Query: 374 GDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFGGKS-----DGNSSPSMKKPYIPNYK 428
N+ + P +LP E L F L++ L S G ++ S K P N+K
Sbjct: 436 TKVFVDNLRVISPKILPTRELLRFLLVLLFNKLKIATSSSKSYSGGATKSTKSPL--NFK 609
Query: 429 LAFEHFCVH 437
+HFC+H
Sbjct: 610 TGVDHFCLH 636
>BI272149 weakly similar to GP|9714501|emb| putative fatty acid elongase {Zea
mays}, partial (15%)
Length = 631
Score = 111 bits (278), Expect(2) = 4e-29
Identities = 63/175 (36%), Positives = 103/175 (58%)
Frame = +2
Query: 53 LGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLASVLASFAVFVFT 112
L + YLI++ + + + LL+V+ ++ VG D E + + ++ SF +
Sbjct: 26 LKHFYLISNIMNILFMSLLIVIATSYVGCFYVND------EYCNPQPSILIKSFCLATSL 187
Query: 113 LSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEESLAFQKRILMSSGI 172
+ FM++P IYL+DFAC++P++ ++E +E A+ G +EES +IL SG+
Sbjct: 188 VYYIFMTRPNKIYLVDFACHKPKNNCLCTKEMLLERANNFGFLNEESFILIHKILERSGV 367
Query: 173 GDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKDIGILVVN 227
G TY+P+A++ T+ E R E++ V+FGA+DEL EKT + KDIGILVVN
Sbjct: 368 GPTTYVPEALLEMPPR-LTLDEARNESNLVLFGAVDELLEKTKVEAKDIGILVVN 529
Score = 35.0 bits (79), Expect(2) = 4e-29
Identities = 13/19 (68%), Positives = 17/19 (89%)
Frame = +3
Query: 230 IFNPTPSLSAMIINHYKMR 248
+FNPTPSLS I+NHYK++
Sbjct: 537 LFNPTPSLSDTIVNHYKLK 593
Score = 29.6 bits (65), Expect = 3.1
Identities = 19/39 (48%), Positives = 23/39 (58%)
Frame = +1
Query: 222 GILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMG 260
G LVV + + L +IIN +RGNIL YNL GMG
Sbjct: 523 GELVVYLILHHHFQILLLIIIN---LRGNILIYNLXGMG 630
>BE203641 weakly similar to PIR|F86141|F861 protein T25K16.11 [imported] -
Arabidopsis thaliana, partial (4%)
Length = 403
Score = 69.3 bits (168), Expect = 3e-12
Identities = 34/96 (35%), Positives = 56/96 (57%)
Frame = +1
Query: 96 SYDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKF 155
S ++ S+L + L+ Y + IYL+DF+CY+P S+E +IE + G F
Sbjct: 85 SSNIFSLLLWCTLIASILTFYLKKCSKNIYLVDFSCYKPFPRSICSKELFIETSRCGGHF 264
Query: 156 DEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTAT 191
+ES+ FQK+I+ SG GD+TY+P+ +++ N T
Sbjct: 265 RDESIDFQKKIMEKSGFGDKTYVPENLLNIPPNICT 372
>CB891954 similar to PIR|A84663|A846 probable beta-ketoacyl-CoA synthase
[imported] - Arabidopsis thaliana, partial (15%)
Length = 402
Score = 62.0 bits (149), Expect(2) = 8e-11
Identities = 33/61 (54%), Positives = 42/61 (68%)
Frame = +1
Query: 151 KSGKFDEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDEL 210
KS K L FQ++IL SG+G+ TY+P+AV+S N +MKE R EA VMFGA+DEL
Sbjct: 178 KSVKLKYVKLDFQRKILERSGLGENTYLPEAVLSIPPN-PSMKEARKEAEAVMFGAIDEL 354
Query: 211 F 211
F
Sbjct: 355 F 357
Score = 28.9 bits (63), Expect = 5.2
Identities = 13/19 (68%), Positives = 15/19 (78%)
Frame = +1
Query: 37 RRLPDFLQSVNLKYVKLGY 55
R LPDF +SV LKYVKL +
Sbjct: 157 RILPDFKKSVKLKYVKLDF 213
Score = 22.7 bits (47), Expect(2) = 8e-11
Identities = 8/12 (66%), Positives = 10/12 (82%)
Frame = +2
Query: 211 FEKTGIRPKDIG 222
F KT ++PKDIG
Sbjct: 356 FSKTTVKPKDIG 391
>BI265034 similar to GP|21536949|gb putative fatty acid elongase {Arabidopsis
thaliana}, partial (23%)
Length = 427
Score = 53.1 bits (126), Expect = 3e-07
Identities = 26/125 (20%), Positives = 59/125 (46%)
Frame = +3
Query: 99 LASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEE 158
L V+ + + +Y + + Y++D+ CY+P + + E ++ ++ +
Sbjct: 51 LLYVVPALYICFLIWKLYDQKRDQECYILDYQCYKPTQDRMLGTEFCGKIIRRTKNLGLD 230
Query: 159 SLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRP 218
F + ++SSGIG++TY P+ V ++ T+ +G E ++ +L ++ I P
Sbjct: 231 EYKFLLKAVVSSGIGEQTYAPRNVFEGRESSPTLNDGIXEMEEFFNDSIAKLLXRSAISP 410
Query: 219 KDIGI 223
+ I
Sbjct: 411 XEXDI 425
>TC79835 homologue to SP|P51082|CHSB_PEA Chalcone synthase 1B (EC 2.3.1.74)
(Naringenin-chalcone synthase 1B). [Garden pea] {Pisum
sativum}, complete
Length = 1393
Score = 38.1 bits (87), Expect = 0.009
Identities = 25/85 (29%), Positives = 41/85 (47%), Gaps = 5/85 (5%)
Frame = +2
Query: 424 IPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYM 483
I +Y F + H ILD+++ L L + M+A+R L +GN SS+ + + L M
Sbjct: 995 ISDYNSIF--WIAHPGGPAILDQVEAKLSLKPEKMQATRHVLSEYGNMSSACVLFILDEM 1168
Query: 484 EAKER----VRRGDRV-WQLAFGSG 503
K + G+ + W + FG G
Sbjct: 1169 RRKSKEDGLATTGEGLEWGVLFGFG 1243
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,786,804
Number of Sequences: 36976
Number of extensions: 267889
Number of successful extensions: 1428
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 1397
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1404
length of query: 538
length of database: 9,014,727
effective HSP length: 101
effective length of query: 437
effective length of database: 5,280,151
effective search space: 2307425987
effective search space used: 2307425987
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0133.9


