

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
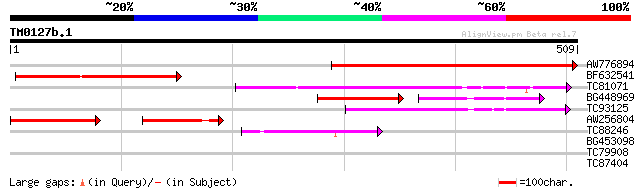
Query= TM0127b.1
(509 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW776894 similar to GP|11762220|gb At1g64860 {Arabidopsis thalia... 399 e-111
BF632541 similar to GP|6682869|dbj| sigma factor {Nicotiana taba... 200 1e-51
TC81071 weakly similar to PIR|T52653|T52653 probable sigma-like ... 138 6e-33
BG448969 similar to PIR|T51715|T517 sigma-like factor [imported]... 87 6e-28
TC93125 similar to PIR|T00707|T00707 transcription initiation fa... 103 2e-22
AW256804 weakly similar to GP|11762220|gb| At1g64860 {Arabidopsi... 92 4e-19
TC88246 similar to PIR|T51328|T51328 transcription initiation fa... 69 6e-12
BG453098 similar to PIR|T51328|T51 transcription initiation fact... 40 0.002
TC79908 similar to GP|14326543|gb|AAK60316.1 AT4g18020/T6K21_200... 35 0.052
TC87404 similar to GP|22137130|gb|AAM91410.1 At1g47330/T3F24_2 {... 29 4.9
>AW776894 similar to GP|11762220|gb At1g64860 {Arabidopsis thaliana}, partial
(43%)
Length = 685
Score = 399 bits (1025), Expect = e-111
Identities = 198/220 (90%), Positives = 210/220 (95%)
Frame = +1
Query: 290 AEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHE 349
AE+ DLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLP HLHE
Sbjct: 13 AELGDLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPAHLHE 192
Query: 350 RLSLIRTAKFRLEERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLP 409
RLSLIR A+++LEERGITPTIDRIAKSLNMSQ+KVRNATEA SKVFSLDREAFPSLNGLP
Sbjct: 193 RLSLIRNAEYKLEERGITPTIDRIAKSLNMSQRKVRNATEATSKVFSLDREAFPSLNGLP 372
Query: 410 GDTHHCYIADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWE 469
GDTHH +IAD + ENIPW+ VDEW LK+EVN+LIN+TLVEREREIIRLYYGLDKECLTWE
Sbjct: 373 GDTHHSFIADKQNENIPWNVVDEWALKEEVNRLINLTLVEREREIIRLYYGLDKECLTWE 552
Query: 470 DISKRIGLSRERVRQVGLVALEKLKHAARKREMEAMLLKH 509
DISKRIGLSRERVRQVGLVALEKLKHAARKREME MLLK+
Sbjct: 553 DISKRIGLSRERVRQVGLVALEKLKHAARKREMETMLLKY 672
>BF632541 similar to GP|6682869|dbj| sigma factor {Nicotiana tabacum},
partial (16%)
Length = 573
Score = 200 bits (508), Expect = 1e-51
Identities = 107/150 (71%), Positives = 124/150 (82%), Gaps = 1/150 (0%)
Frame = +2
Query: 6 VIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGSTHYQIVPAKSVTVAKKSSDYTQAFPASD 65
VIGLSGGKRLLSSSY+YSDIIEK S + DFGST YQ+ P+KSV VAKKSS YT FP +
Sbjct: 62 VIGLSGGKRLLSSSYHYSDIIEKFSHACDFGSTQYQLPPSKSVIVAKKSSKYTPTFPP-E 238
Query: 66 RPNQSIKALKEHV-DGVPAAAETWFQECDSNDLEVESSDLGYSVEALLLLQKSMLEKQWS 124
R NQSIKALKEHV D A +E WFQ +SND EVESSD+ YS+++LLLLQKSMLEKQWS
Sbjct: 239 RKNQSIKALKEHVVDSAQADSEQWFQGYNSNDFEVESSDMDYSLDSLLLLQKSMLEKQWS 418
Query: 125 LSCEREVLTEHPRQEKSSKKVAVTCSGVSA 154
LS ER+VL+E+ R+EK +KV+VTCSGVSA
Sbjct: 419 LSFERKVLSENSRREKIRRKVSVTCSGVSA 508
>TC81071 weakly similar to PIR|T52653|T52653 probable sigma-like transcription
initiation factor [imported] - Arabidopsis thaliana
(fragment), partial (64%)
Length = 1856
Score = 138 bits (347), Expect = 6e-33
Identities = 98/307 (31%), Positives = 171/307 (54%), Gaps = 5/307 (1%)
Frame = +3
Query: 203 SEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISRSELRAKTIE 262
++QLLS E +L +I+ L E K +L+ G EP+ + A + + +L+ K +
Sbjct: 801 TKQLLSIEEESQLIAQIQDLFKLKETKIKLQSHFGREPTLAEWADGVGLICRDLQ-KRLH 977
Query: 263 C-TLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKGFKISTY 321
C ++++L +N+RLV+ IA+ Y G + DL+Q G +GL+R +EKF G + STY
Sbjct: 978 CGNRSKDKLIHANLRLVVHIAKYYQGRGLSLQDLLQEGSMGLMRSVEKFKPQAGCRFSTY 1157
Query: 322 VYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTA-KFRLEERGITPTIDRIAKSLNMS 380
YWWIRQ + RAL +S+T+RLP + + L I A K ++E + PT + IAK + ++
Sbjct: 1158 AYWWIRQTIRRALFLHSKTIRLPENFYALLGKIAEAKKSYIKEGNLHPTKEEIAKRVGIT 1337
Query: 381 QKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIADNRLENIPWHGVDEWTLKDEVN 440
++ + S++R + + T AD+ +E IP V + ++ V
Sbjct: 1338 VDRLEMLLFSTRTPLSMERAVWADSD----TTFQEITADSSIE-IPNVCVAKQLMRSHVR 1502
Query: 441 KLINVTLVEREREIIRLYYGLD---KECLTWEDISKRIGLSRERVRQVGLVALEKLKHAA 497
L+ + L +ER+++RL +G++ ++ LT +I K +G+ +ERVRQ+ AL KLK
Sbjct: 1503 NLLGI-LPPKERKVLRLRFGIEDGYEKSLT--EIGKVLGVCKERVRQLESQALNKLKQCL 1673
Query: 498 RKREMEA 504
++++A
Sbjct: 1674 VSQQLDA 1694
>BG448969 similar to PIR|T51715|T517 sigma-like factor [imported] -
Arabidopsis thaliana, partial (22%)
Length = 617
Score = 87.4 bits (215), Expect(2) = 6e-28
Identities = 40/77 (51%), Positives = 53/77 (67%)
Frame = +3
Query: 277 LVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVE 336
LV SIA Y G + DL+Q G IGLL G EKFD ++G K+STYVYWWI+QG+ +AL +
Sbjct: 3 LVASIAAHYQGKGLSIQDLIQEGTIGLLHGAEKFDPNRGCKLSTYVYWWIKQGIIKALAK 182
Query: 337 NSRTLRLPTHLHERLSL 353
SR +RLP ++ S+
Sbjct: 183 KSRLIRLPFVIYREKSM 233
Score = 55.1 bits (131), Expect(2) = 6e-28
Identities = 42/114 (36%), Positives = 63/114 (54%), Gaps = 1/114 (0%)
Frame = +2
Query: 368 PTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIADNRLENIPW 427
PT + IA+ LN++ V+ +E + SLDR N + + I +E IP
Sbjct: 293 PTYNEIAEVLNVNVSTVKLVSERSRQPISLDRSITDQSNLILKE-----IIPGPVEMIPE 457
Query: 428 HGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKEC-LTWEDISKRIGLSRE 480
V+ +K V KL+N TL +RE EI+RL +GL+ E L++E+I K + LSRE
Sbjct: 458 KMVERQLMKQGVVKLLN-TLDKREEEIVRLXFGLNGETPLSFEEIGKLLKLSRE 616
>TC93125 similar to PIR|T00707|T00707 transcription initiation factor sigma
homolog [imported] - Arabidopsis thaliana, partial (35%)
Length = 656
Score = 103 bits (256), Expect = 2e-22
Identities = 63/204 (30%), Positives = 117/204 (56%), Gaps = 2/204 (0%)
Frame = +2
Query: 302 GLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTAKFRL 361
GL+RG EK+D+SKGFK STY +WWI+Q V ++L SRT+RLP H+ ++ A+ +L
Sbjct: 2 GLVRGAEKYDASKGFKFSTYAHWWIKQAVRKSLSVQSRTIRLPFHMVGATYKVKEARKQL 181
Query: 362 -EERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIADN 420
E G P + +A++ +S K++ SL+++ + + + +AD
Sbjct: 182 YSENGRQPDDEEVAEATGLSMKRLSAVLLTPKAPRSLEQKVGINQSLKLSEV----LADP 349
Query: 421 RLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDK-ECLTWEDISKRIGLSR 479
E + ++ +K ++ K+++ +L RE+++I+ +G+D T ++I + +G+SR
Sbjct: 350 EAETAEEQLIKQF-MKKDLEKVLD-SLNPREKQVIKWRFGMDDGRMKTLQEIGEMMGVSR 523
Query: 480 ERVRQVGLVALEKLKHAARKREME 503
ER+RQ+ A KLK+ R + ++
Sbjct: 524 ERIRQIESCAFRKLKNKKRAKHLQ 595
>AW256804 weakly similar to GP|11762220|gb| At1g64860 {Arabidopsis thaliana},
partial (10%)
Length = 428
Score = 92.4 bits (228), Expect = 4e-19
Identities = 47/73 (64%), Positives = 60/73 (81%)
Frame = +2
Query: 120 EKQWSLSCEREVLTEHPRQEKSSKKVAVTCSGVSARQRRINTKRKIPGKTGSAMQACDAM 179
+KQWSLS ER+VL+E+ R+EK +KV+VTCSGVSARQRR+ TKRK+ K+GSA+ M
Sbjct: 224 QKQWSLSFERKVLSENSRREKIRRKVSVTCSGVSARQRRMTTKRKVAAKSGSAV-----M 388
Query: 180 QMRSLISPELLQN 192
Q+RS ISPEL+QN
Sbjct: 389 QLRSTISPELIQN 427
Score = 90.1 bits (222), Expect = 2e-18
Identities = 48/81 (59%), Positives = 59/81 (72%)
Frame = +1
Query: 1 MATAAVIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGSTHYQIVPAKSVTVAKKSSDYTQA 60
MATAAVIGLSGGKRLLSSSY+YSDIIEK S + DFGST YQ+ P+KSV VAKKSS ++
Sbjct: 61 MATAAVIGLSGGKRLLSSSYHYSDIIEKFSHACDFGSTQYQLPPSKSVIVAKKSSKAMES 240
Query: 61 FPASDRPNQSIKALKEHVDGV 81
F + + K+ +G+
Sbjct: 241 FV*KESVE*EFEERKDPEEGI 303
>TC88246 similar to PIR|T51328|T51328 transcription initiation factor sigma5
plastid -specific [imported] - Arabidopsis thaliana,
partial (42%)
Length = 1158
Score = 68.6 bits (166), Expect = 6e-12
Identities = 40/129 (31%), Positives = 74/129 (57%), Gaps = 3/129 (2%)
Frame = +3
Query: 209 HAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISRSELRAKTIECTLARE 268
HA + KL + +K L + K L+++L EP++D+IA + + ++++ AR
Sbjct: 768 HAFLFKLMQPMKALLQV---KDDLQKELEREPTEDEIADATNMHTTQVKKAIEVGRAARN 938
Query: 269 RLAMSNVRLVMSIAQRYDNMGAE---MADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWW 325
+L N+RLV+ + RY + A DL Q G+ GL+ I++F+ ++ F++STY +W
Sbjct: 939 KLIKHNLRLVLFVINRYFSDFANSPRFQDLCQAGVKGLITSIDRFEPNRRFRLSTYSLFW 1118
Query: 326 IRQGVSRAL 334
IR + R++
Sbjct: 1119IRHAIIRSM 1145
>BG453098 similar to PIR|T51328|T51 transcription initiation factor sigma5
plastid -specific [imported] - Arabidopsis thaliana,
partial (23%)
Length = 659
Score = 40.0 bits (92), Expect = 0.002
Identities = 38/137 (27%), Positives = 70/137 (50%), Gaps = 5/137 (3%)
Frame = -3
Query: 367 TPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPS----LNGLPGDTHHCYIADNRL 422
+PT + I + +++S ++ R+ +A + SL + +NG+ D DNR
Sbjct: 636 SPTEEEIIEKVHVSPERYRDVMKASKPLLSLHSRHLTTQEEFINGVVDDGG--VDGDNRR 463
Query: 423 ENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLD-KECLTWEDISKRIGLSRER 481
+ L+ ++ +++ +L +E +IR +GLD K T +I+ + +SRE
Sbjct: 462 QPA--------LLRLALDDVLD-SLKPKENLVIRQRFGLDGKGDRTLGEIASNLNISREM 310
Query: 482 VRQVGLVALEKLKHAAR 498
VR+ + AL KLKH+AR
Sbjct: 309 VRKHEVKALMKLKHSAR 259
>TC79908 similar to GP|14326543|gb|AAK60316.1 AT4g18020/T6K21_200 {Arabidopsis
thaliana}, partial (44%)
Length = 1599
Score = 35.4 bits (80), Expect = 0.052
Identities = 36/134 (26%), Positives = 61/134 (44%), Gaps = 7/134 (5%)
Frame = +2
Query: 113 LLQKSMLEKQWSLSCEREVLTEHPRQEKSSKKVAVTCSGVSARQRRINTKRKIPGKTGSA 172
LL ++Q + S E+E EH + SK V +TC ++A TK +
Sbjct: 890 LLDDGDCQEQTNCSTEKES-GEH---DGESKSVEITCENLNAESITQQTKSEATLVQKEE 1057
Query: 173 MQACDAMQMRSLISPEL----LQNRSKGYVKGVVSEQLLSH-AEVVKLSEKIKVGLSLDE 227
+ DA + S++SP+L + N S+ + SH E+ +K+KV + +
Sbjct: 1058 EEFADASKCESVVSPQLPNVKVLNNSESNTTSANKVVVRSHKCEIKANRKKMKVDWTPEL 1237
Query: 228 HKSRLK--EKLGCE 239
HK +K E+LG +
Sbjct: 1238 HKKFVKAVEQLGID 1279
>TC87404 similar to GP|22137130|gb|AAM91410.1 At1g47330/T3F24_2 {Arabidopsis
thaliana}, partial (70%)
Length = 1944
Score = 28.9 bits (63), Expect = 4.9
Identities = 15/36 (41%), Positives = 23/36 (63%)
Frame = +3
Query: 373 IAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGL 408
IA +L +++K ++A ISK FSLD +A +L L
Sbjct: 681 IAGALELTEKTAKDAMTPISKAFSLDLDATLNLETL 788
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.131 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,935,313
Number of Sequences: 36976
Number of extensions: 171502
Number of successful extensions: 865
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 857
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 860
length of query: 509
length of database: 9,014,727
effective HSP length: 100
effective length of query: 409
effective length of database: 5,317,127
effective search space: 2174704943
effective search space used: 2174704943
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0127b.1


