

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
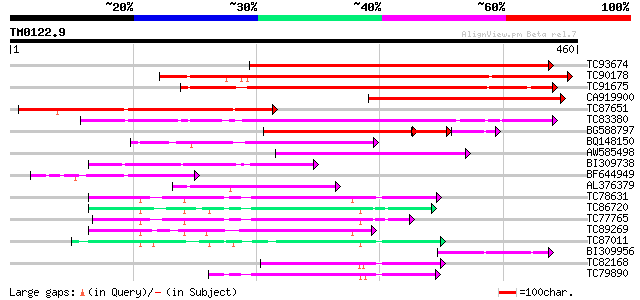
Query= TM0122.9
(460 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC93674 similar to GP|15292729|gb|AAK92733.1 putative polygalact... 358 2e-99
TC90178 similar to GP|13958032|gb|AAK50769.1 polygalacturonase {... 331 3e-91
TC91675 similar to GP|6624205|dbj|BAA88472.1 polygalacturonase {... 248 3e-66
CA919900 similar to GP|15292729|gb putative polygalacturonase PG... 234 5e-62
TC87651 similar to GP|15292729|gb|AAK92733.1 putative polygalact... 224 4e-59
TC83380 weakly similar to GP|10176984|dbj|BAB10216. polygalactur... 208 3e-54
BG588797 weakly similar to PIR|T08215|T08 polygalacturonase (EC ... 135 2e-42
BQ148150 weakly similar to GP|10185719|gb NTS1 protein {Nicotian... 142 4e-34
AW585498 weakly similar to GP|10177065|db polygalacturonase-like... 139 2e-33
BI309738 weakly similar to GP|15028105|gb putative polygalacturo... 127 7e-30
BF644949 similar to GP|15292729|gb putative polygalacturonase PG... 122 4e-28
AL376379 homologue to GP|13958032|gb polygalacturonase {Pisum sa... 99 3e-21
TC78631 similar to PIR|T05388|T05388 hypothetical protein F16G20... 88 8e-18
TC86720 similar to PIR|T47941|T47941 hypothetical protein F2A19.... 86 4e-17
TC77765 similar to PIR|T47941|T47941 hypothetical protein F2A19.... 83 3e-16
TC89269 similar to PIR|T05614|T05614 hypothetical protein F9D16.... 79 4e-15
TC87011 weakly similar to GP|22136814|gb|AAM91751.1 unknown prot... 77 2e-14
BI309956 weakly similar to GP|7381227|gb|A polygalacturonase {Ly... 77 2e-14
TC82168 weakly similar to GP|11762132|gb|AAG40344.1 AT3g62110 {A... 75 7e-14
TC79890 similar to GP|11762132|gb|AAG40344.1 AT3g62110 {Arabidop... 71 8e-13
>TC93674 similar to GP|15292729|gb|AAK92733.1 putative polygalacturonase PG1
{Arabidopsis thaliana}, partial (51%)
Length = 745
Score = 358 bits (920), Expect = 2e-99
Identities = 162/247 (65%), Positives = 208/247 (83%)
Frame = +3
Query: 195 SPVAIRFFMSSNLTVQGLRVKNSPQFNFRFDGCIKVHVESIYITAPKLSPNTDGIHIENT 254
SP IRFFMSSNL ++GL+++NSPQF+ +FDGC V ++ + I+APKLSPNTDGIH+ NT
Sbjct: 3 SPTMIRFFMSSNLVLRGLKIQNSPQFHVKFDGCQGVLIDELSISAPKLSPNTDGIHLGNT 182
Query: 255 NDVKIYNSMISNGDDCVSIGSGCYDVDIKNITCGPGHGISIGSLGIHNSRACVSNITVRD 314
DV IYNS+ISNGDDC+SIG GC +V++ +TC P HGISIGSLG+HNS ACVSN+TVR+
Sbjct: 183 RDVGIYNSLISNGDDCISIGPGCSNVNVDGVTCAPTHGISIGSLGVHNSHACVSNLTVRN 362
Query: 315 SMMKGTDNGVRIKTWQGGSGSVSGVKFSNIRMVNVRNPIIIDQFYCITEGCTNKTSAVFV 374
S++K +DNG+RIKTWQGG+GSV+G+ F NI+M NVRN I IDQFYC+++ C N+TSAV+V
Sbjct: 363 SIIKESDNGLRIKTWQGGTGSVTGLTFDNIQMENVRNCINIDQFYCLSKECMNQTSAVYV 542
Query: 375 SNILYTNIKGTYDVRSLPMRFACSDSIPCTHLALSDIELLPAEGDIAHDPFCWNAYGDLQ 434
+NI Y IKGTYDVR+ P+ FACSD++ CT++ LS+IELLP EG++ DPFCWNAYG +
Sbjct: 543 NNISYRKIKGTYDVRTPPIHFACSDTVACTNITLSEIELLPYEGELVDDPFCWNAYGRQE 722
Query: 435 TLTIPPV 441
TLTIPP+
Sbjct: 723 TLTIPPL 743
>TC90178 similar to GP|13958032|gb|AAK50769.1 polygalacturonase {Pisum
sativum}, partial (94%)
Length = 1114
Score = 331 bits (849), Expect = 3e-91
Identities = 169/356 (47%), Positives = 227/356 (63%), Gaps = 21/356 (5%)
Frame = +3
Query: 122 IVFKVEGTLMPPDGPESWPKNISRHQWLVFYRINGMSLEGSGLIDGRGEKWWD------- 174
I+F+V+GT++ P +W + + QWL F ++ G +++G+G+ DGRG WW
Sbjct: 12 IIFQVDGTIIAPTNSNAWGRGLL--QWLDFTKLVGFTIQGNGIFDGRGSVWWQDTQYNDP 185
Query: 175 --------LPCKPHRQGPHW---TTLPG--PCDSPVAIRFFMSSNLTVQGLRVKNSPQFN 221
+P P ++L G P P AIRF+ S N TV G+ ++NSPQ +
Sbjct: 186 LDDEEKLLVPLNNTIVSPPMQIESSLGGKMPSIKPTAIRFYGSINPTVTGITIQNSPQCH 365
Query: 222 FRFDGCIKVHVESIYITAPKLSPNTDGIHIENTNDVKIYNSMISNGDDCVSIGSGCYDVD 281
+FD C V V + I++P SPNTDGIH++N+ DV IY S ++ GDDC+SI +GC +V
Sbjct: 366 LKFDNCNGVLVHDVTISSPGDSPNTDGIHLQNSRDVLIYKSTMACGDDCISIQTGCSNVY 545
Query: 282 IKNITCGPGHGISIGSLGIHNSRACVSNITVRDSMMKGTDNGVRIKTWQGGSGSVSGVKF 341
+ N+ CGPGHGISIG LG N+RACVSNITVRD M T NGVRIKTWQGGSGSV GV F
Sbjct: 546 VHNVDCGPGHGISIGGLGKDNTRACVSNITVRDVNMHNTMNGVRIKTWQGGSGSVQGVLF 725
Query: 342 SNIRMVNVRNPIIIDQFYCITEGCTNKTSAVFVSNILYTNIKGTYDVRSLPMRFACSDSI 401
SNI++ V+ PIIIDQFYC C N+T+AV ++ I Y IKGTY V+ P+ FACSDS+
Sbjct: 726 SNIQVSQVQVPIIIDQFYCDKRNCKNQTAAVALTGINYEGIKGTYTVK--PVHFACSDSL 899
Query: 402 PCTHLALSDIELLPAEGDI-AHDPFCWNAYGDLQTLTIPPVSCLMQGTPQSWLDHD 456
PC ++L+ +EL P + +DPFCW YG+L+T T+PP+ CL D D
Sbjct: 900 PCIDVSLTSVELQPVQDKYHLYDPFCWETYGELKTATVPPIDCLQNWETTEQSDSD 1067
>TC91675 similar to GP|6624205|dbj|BAA88472.1 polygalacturonase {Cucumis
sativus}, partial (65%)
Length = 1146
Score = 248 bits (634), Expect = 3e-66
Identities = 125/308 (40%), Positives = 189/308 (60%), Gaps = 2/308 (0%)
Frame = +1
Query: 139 WPKNISRHQWLVFYRINGMSLEGSGLIDGRGEKWWDLPCKPHRQGPHWTTLPGPCD-SPV 197
W + R WL F ++ +GSG+IDG G KWW CK ++ P C +P
Sbjct: 1 WDPKLPR-VWLDFSKLKKAVFQGSGVIDGSGSKWWAASCKKNKTNP--------CKGAPT 153
Query: 198 AIRFFMSSNLTVQGLRVKNSPQFNFRFDGCIKVHVESIYITAPKLSPNTDGIHIENTNDV 257
A+ SS++ V+GL ++N+ Q +F C V + + + +P SPNTDGIHI + +V
Sbjct: 154 ALTIDTSSSIKVKGLTIQNNQQMHFTISRCDSVRILGVKVASPGDSPNTDGIHISESTNV 333
Query: 258 KIYNSMISNGDDCVSIGSGCYDVDIKNITCGPGHGISIGSLGIHNSRACVSNITVRDSMM 317
+ + I GDDC+SI + ++ +K I CGPGHGISIGSLG NS V+ + + + +
Sbjct: 334 IVQDCKIGTGDDCISIVNASSNIKMKRIFCGPGHGISIGSLGKDNSTGVVTKVILDTAFL 513
Query: 318 KGTDNGVRIKTWQGGSGSVSGVKFSNIRMVNVRNPIIIDQFYCIT-EGCTNKTSAVFVSN 376
KGT NG+RIKTWQGG+G V GV+F N+R+ NV NPI+IDQFYC + C N++SA+ +S
Sbjct: 514 KGTTNGLRIKTWQGGAGYVKGVRFQNVRVENVSNPILIDQFYCDSPTSCQNQSSALEISE 693
Query: 377 ILYTNIKGTYDVRSLPMRFACSDSIPCTHLALSDIELLPAEGDIAHDPFCWNAYGDLQTL 436
I+Y N+ GT + + ++F CSD++PC++L LS+++L +G + + +C +A G +
Sbjct: 694 IMYQNVSGTTN-SAKAIKFDCSDTVPCSNLVLSNVDLEKQDGTV--ETYCHSAQGFGYGV 864
Query: 437 TIPPVSCL 444
P CL
Sbjct: 865 VHPSAECL 888
>CA919900 similar to GP|15292729|gb putative polygalacturonase PG1
{Arabidopsis thaliana}, partial (33%)
Length = 752
Score = 234 bits (597), Expect = 5e-62
Identities = 102/160 (63%), Positives = 137/160 (84%)
Frame = -2
Query: 292 GISIGSLGIHNSRACVSNITVRDSMMKGTDNGVRIKTWQGGSGSVSGVKFSNIRMVNVRN 351
GISIGSLG+HNS+ACVSN+TVRD++++ +DNG+RIKTWQGG GSVS +KF NI+M NV N
Sbjct: 751 GISIGSLGVHNSQACVSNLTVRDTVIRESDNGLRIKTWQGGMGSVSNLKFENIQMENVGN 572
Query: 352 PIIIDQFYCITEGCTNKTSAVFVSNILYTNIKGTYDVRSLPMRFACSDSIPCTHLALSDI 411
I+IDQ+YC+T+ C N+TSAV V+++ Y NIKGTYDVR+ P+ FACSD++ CT++ LS++
Sbjct: 571 CILIDQYYCLTKECLNQTSAVHVNDVSYKNIKGTYDVRTAPIHFACSDTVACTNITLSEV 392
Query: 412 ELLPAEGDIAHDPFCWNAYGDLQTLTIPPVSCLMQGTPQS 451
ELLP EG++ DPFCWNAYG +TLTIPP++CL +G P++
Sbjct: 391 ELLPFEGELLDDPFCWNAYGAQETLTIPPINCLREGDPET 272
>TC87651 similar to GP|15292729|gb|AAK92733.1 putative polygalacturonase PG1
{Arabidopsis thaliana}, partial (35%)
Length = 822
Score = 224 bits (572), Expect = 4e-59
Identities = 111/215 (51%), Positives = 146/215 (67%), Gaps = 5/215 (2%)
Frame = +3
Query: 8 HILPLAFCIFSLTLFLSAQARHHSHTKHNK-----HSHSHKSTKTSPPPPPHNCYNATGI 62
H L + + F L + R+H HTK K S+S S + P P ++ I
Sbjct: 186 HGLFILWITFILLNLRNVDGRNHLHTKQKKVSSLLPSNSSPSVPSDPYPNDPRDSSSDCI 365
Query: 63 FDVTTFGAVGDGIADDTVSFKMAWDTACQSELPGNVILVPKGFSFIIQSTIFTGPCQGGI 122
FDVT+FGA+GDG ADDT +FK AW AC E ++LVP+ ++F+I S IF+GPC+ G+
Sbjct: 366 FDVTSFGAIGDGSADDTPAFKKAWKAACAVE--SGILLVPENYTFMITSIIFSGPCKPGL 539
Query: 123 VFKVEGTLMPPDGPESWPKNISRHQWLVFYRINGMSLEGSGLIDGRGEKWWDLPCKPHRQ 182
VF+V+G LM PDGP+SWP+ SR+QWLVFY+++ MSL G+G I+G G++WWDLPCKPHR
Sbjct: 540 VFQVDGMLMAPDGPDSWPEADSRNQWLVFYKLDQMSLNGTGTIEGNGDQWWDLPCKPHR- 716
Query: 183 GPHWTTLPGPCDSPVAIRFFMSSNLTVQGLRVKNS 217
GP TL GPC SP IRFFMSSNL V GL+++NS
Sbjct: 717 GPDGKTLSGPCGSPAMIRFFMSSNLYVNGLKIENS 821
>TC83380 weakly similar to GP|10176984|dbj|BAB10216. polygalacturonase-like
protein {Arabidopsis thaliana}, partial (21%)
Length = 1220
Score = 208 bits (530), Expect = 3e-54
Identities = 129/389 (33%), Positives = 198/389 (50%), Gaps = 2/389 (0%)
Frame = +2
Query: 58 NATGIFDVTTFGAVGDGIADDTVSFKMAWDTACQSELPGNVILVPKGFSFIIQSTI-FTG 116
N +V +GA GDG DD+ +F A+ C GN ++VP G SF ++ T+ F+G
Sbjct: 77 NGEDTLNVIHYGAKGDGRTDDSKAFVDAFKALCGGRY-GNTLVVPNGHSFFVRPTLNFSG 253
Query: 117 PCQG-GIVFKVEGTLMPPDGPESWPKNISRHQWLVFYRINGMSLEGSGLIDGRGEKWWDL 175
PC I K+ G ++ P + W + S WL F+ I+G++L+GSG+I+G GE W
Sbjct: 254 PCYSKNINIKIMGNILAPKRSD-WGRECSL-MWLHFFNISGLTLDGSGVINGNGEGW--- 418
Query: 176 PCKPHRQGPHWTTLPGPCDSPVAIRFFMSSNLTVQGLRVKNSPQFNFRFDGCIKVHVESI 235
+ +G + G P A++F L + GL N P + + + I
Sbjct: 419 --ESREKG-----IGGCPRIPTALQFDKCDGLQINGLTHINGPGAHIAVIDSQDITISHI 577
Query: 236 YITAPKLSPNTDGIHIENTNDVKIYNSMISNGDDCVSIGSGCYDVDIKNITCGPGHGISI 295
+I +PK S NTDGI + T V I++ I +GDDC+++ G V++ N+TCGPGHGIS+
Sbjct: 578 HINSPKKSHNTDGIDLTRTIGVNIHDIQIESGDDCIAVKGGSQFVNVSNVTCGPGHGISV 757
Query: 296 GSLGIHNSRACVSNITVRDSMMKGTDNGVRIKTWQGGSGSVSGVKFSNIRMVNVRNPIII 355
GSLG H S V + +V++ G D+ V+IKTW GG G + F +I + P+ I
Sbjct: 758 GSLGGHGSEEFVQHFSVKNCTFNGADSAVKIKTWPGGKGYAKHIIFEDIIINQTNYPVFI 937
Query: 356 DQFYCITEGCTNKTSAVFVSNILYTNIKGTYDVRSLPMRFACSDSIPCTHLALSDIELLP 415
DQ Y T + AV +SNI ++NI GT + + C+ I C ++ L+ I +
Sbjct: 938 DQHYMRT---PEQHQAVKISNITFSNIYGTC-IGEDAVVLDCA-KIGCYNITLNQINITS 1102
Query: 416 AEGDIAHDPFCWNAYGDLQTLTIPPVSCL 444
C N +G + P C+
Sbjct: 1103INRKKPASVKCKNVHGTANDIIAPHGLCV 1189
>BG588797 weakly similar to PIR|T08215|T08 polygalacturonase (EC 3.2.1.15) 3
precursor - muskmelon, partial (35%)
Length = 631
Score = 135 bits (340), Expect(3) = 2e-42
Identities = 67/125 (53%), Positives = 84/125 (66%)
Frame = +2
Query: 207 LTVQGLRVKNSPQFNFRFDGCIKVHVESIYITAPKLSPNTDGIHIENTNDVKIYNSMISN 266
L V+ LR K+S Q + F+ C V V ++ + AP+ SPNTDGIH+ T ++ I N I
Sbjct: 2 LKVKNLRFKDSQQMHVVFERCFNVFVSNLIVRAPEDSPNTDGIHVAETQNIDIINCDIGT 181
Query: 267 GDDCVSIGSGCYDVDIKNITCGPGHGISIGSLGIHNSRACVSNITVRDSMMKGTDNGVRI 326
GDDC+SI SG +V +ITCGPGHGISIGSLG NS A VSN+ V + + GT NGVRI
Sbjct: 182 GDDCISIVSGSKNVRAIDITCGPGHGISIGSLGADNSEAEVSNVVVNRAALTGTTNGVRI 361
Query: 327 KTWQG 331
KTW G
Sbjct: 362 KTWPG 376
Score = 43.1 bits (100), Expect(3) = 2e-42
Identities = 19/32 (59%), Positives = 23/32 (71%)
Frame = +3
Query: 327 KTWQGGSGSVSGVKFSNIRMVNVRNPIIIDQF 358
K QGGSG +KF NI+M NV NP+IIDQ+
Sbjct: 363 KHGQGGSGYARNIKFMNIKMQNVTNPVIIDQY 458
Score = 32.7 bits (73), Expect(3) = 2e-42
Identities = 17/41 (41%), Positives = 24/41 (58%), Gaps = 1/41 (2%)
Frame = +1
Query: 359 YCI-TEGCTNKTSAVFVSNILYTNIKGTYDVRSLPMRFACS 398
YC TE C + AV +S +LY NI+GT + ++F CS
Sbjct: 460 YCDQTEPCQERNKAVQLSQVLYQNIRGT-SASEIAIKFNCS 579
>BQ148150 weakly similar to GP|10185719|gb NTS1 protein {Nicotiana tabacum},
partial (31%)
Length = 669
Score = 142 bits (357), Expect = 4e-34
Identities = 79/206 (38%), Positives = 117/206 (56%), Gaps = 5/206 (2%)
Frame = +2
Query: 99 ILVPKGFSFIIQSTIFTGPCQGGIVFKVEGTLMPPDGPESWPKNISRH---QWLVFYRIN 155
+L+P G +F+ T+F GPC VE G ++S + +W F I+
Sbjct: 11 VLIPAG-TFVTGQTLFAGPCTSPKPITVEIV-----GKAQATTDLSEYYSPEWFQFENID 172
Query: 156 GMSLEGSGLIDGRGEKWWDL-PCKPHRQGPHWTTLPGPCDS-PVAIRFFMSSNLTVQGLR 213
G+ L+GSG+ DG+G W L CK ++ G C S P +++F N VQ +
Sbjct: 173 GLVLKGSGVFDGQGSISWPLNDCKQNK---------GNCASLPSSLKFDKIKNAIVQDVT 325
Query: 214 VKNSPQFNFRFDGCIKVHVESIYITAPKLSPNTDGIHIENTNDVKIYNSMISNGDDCVSI 273
NS QF+F GC + +++ITAP SPNTDG+HI +++ + + NS+I+ GDDC+S+
Sbjct: 326 SLNSMQFHFHLHGCSNISFTNLHITAPGNSPNTDGMHISSSDFITVTNSVIATGDDCISV 505
Query: 274 GSGCYDVDIKNITCGPGHGISIGSLG 299
G ++ I ITCGPGHGIS+GSLG
Sbjct: 506 GHSTSNITISGITCGPGHGISVGSLG 583
>AW585498 weakly similar to GP|10177065|db polygalacturonase-like protein
{Arabidopsis thaliana}, partial (26%)
Length = 583
Score = 139 bits (350), Expect = 2e-33
Identities = 71/160 (44%), Positives = 95/160 (59%), Gaps = 1/160 (0%)
Frame = +3
Query: 216 NSPQFNFRFDGCIKVHVESIYITAPKLSPNTDGIHIENTNDVKIYNSMISNGDDCVSIGS 275
N + + C K + +I + AP SPNTDGI I + D+++ NS I+ GDDC++I +
Sbjct: 15 NPSRSHITLTSCKKGIISNIRLIAPGESPNTDGIDISASRDIQVLNSFIATGDDCIAISA 194
Query: 276 GCYDVDIKNITCGPGHGISIGSLGIHNSRACVSNITVRDSMMKGTDNGVRIKTWQGGSGS 335
G + I ITCGPGHGISIGSLG V ++ V++ + T GVRIKT QGG G
Sbjct: 195 GSSVIKITAITCGPGHGISIGSLGARGDTDIVEDVHVKNCTLTETLTGVRIKTKQGGGGF 374
Query: 336 VSGVKFSNIRMVNVRNPIIIDQFYCITE-GCTNKTSAVFV 374
+ F NI+ V NPI IDQFYC+ + C N T A+ V
Sbjct: 375 ARRITFENIKFVRAHNPIWIDQFYCVNQMVCRNMTKAIKV 494
>BI309738 weakly similar to GP|15028105|gb putative polygalacturonase
{Arabidopsis thaliana}, partial (31%)
Length = 768
Score = 127 bits (320), Expect = 7e-30
Identities = 66/187 (35%), Positives = 103/187 (54%), Gaps = 1/187 (0%)
Frame = +2
Query: 65 VTTFGAVGDGIADDTVSFKMAWDTACQSELPGNV-ILVPKGFSFIIQSTIFTGPCQGGIV 123
+ FGA GDG +DT +F W+ AC L G + I+ P +F++ GPC+ I
Sbjct: 236 IADFGARGDGFHNDTQAFLKVWEIACS--LSGFINIVFPYQKTFLVTPIDIGGPCRSKIT 409
Query: 124 FKVEGTLMPPDGPESWPKNISRHQWLVFYRINGMSLEGSGLIDGRGEKWWDLPCKPHRQG 183
++ G ++ P P+ W +++ +W+ F+ +N +S+EG G IDG G++WW CK +
Sbjct: 410 LRILGAIVAPRNPDVW-HGLNKRKWIYFHGVNHLSVEGGGRIDGMGQEWWSRSCKINTTN 586
Query: 184 PHWTTLPGPCDSPVAIRFFMSSNLTVQGLRVKNSPQFNFRFDGCIKVHVESIYITAPKLS 243
P LP +P A+ F +L V+ L V NS + + F C++V + + AP LS
Sbjct: 587 P---CLP----APTALTFHKCKSLKVRNLTVLNSQKMHIAFTSCMRVVASRLKVLAPALS 745
Query: 244 PNTDGIH 250
PNTDGIH
Sbjct: 746 PNTDGIH 766
>BF644949 similar to GP|15292729|gb putative polygalacturonase PG1
{Arabidopsis thaliana}, partial (22%)
Length = 673
Score = 122 bits (305), Expect = 4e-28
Identities = 67/154 (43%), Positives = 92/154 (59%), Gaps = 17/154 (11%)
Frame = +1
Query: 18 SLTLFL-SAQARHHSHTKHNKHSHSHKSTKTSPPPP----------------PHNCYNAT 60
++ LF+ + + R+H H K K S + + SPP P P NC
Sbjct: 238 TMILFIHNVEGRYH-HKKPKKTSPA--PSDPSPPSPSFPSDPYPYPNDPGESPSNC---- 396
Query: 61 GIFDVTTFGAVGDGIADDTVSFKMAWDTACQSELPGNVILVPKGFSFIIQSTIFTGPCQG 120
+FDV +FGAVGDG ADDT +F+ AW AC + V+L P+ + F I STIF+GPC+
Sbjct: 397 -VFDVRSFGAVGDGDADDTAAFRAAWKAACAVD--SGVLLAPENYCFKITSTIFSGPCKP 567
Query: 121 GIVFKVEGTLMPPDGPESWPKNISRHQWLVFYRI 154
G+VF+++GTLM PDGP WP+ S+ QWLVFYR+
Sbjct: 568 GLVFQIDGTLMAPDGPNCWPEADSKSQWLVFYRL 669
>AL376379 homologue to GP|13958032|gb polygalacturonase {Pisum sativum},
partial (43%)
Length = 459
Score = 99.0 bits (245), Expect = 3e-21
Identities = 55/155 (35%), Positives = 83/155 (53%), Gaps = 19/155 (12%)
Frame = +1
Query: 133 PDGPESWPKNISRHQWLVFYRINGMSLEGSGLIDGRGEKWW-DLPC-------------- 177
P P +W QWL ++ G++++G+G+IDGRG WW D P
Sbjct: 1 PTNPNAWSGVTL--QWLECTKLEGITIQGNGVIDGRGSVWWQDFPYDNPIDDEEKLIVPL 174
Query: 178 ----KPHRQGPHWTTLPGPCDSPVAIRFFMSSNLTVQGLRVKNSPQFNFRFDGCIKVHVE 233
KP + P + P A+RF+ S TV G+ ++NSPQ + +FD C V V
Sbjct: 175 NQTQKPPMPVQNEMGRKMPSNKPTALRFYGSYGPTVTGITIQNSPQCHLKFDNCNGVLVH 354
Query: 234 SIYITAPKLSPNTDGIHIENTNDVKIYNSMISNGD 268
+ I++P SPNTDGIH++N+ DV I++S ++ GD
Sbjct: 355 DVSISSPGDSPNTDGIHLQNSKDVLIHSSKLACGD 459
>TC78631 similar to PIR|T05388|T05388 hypothetical protein F16G20.200 -
Arabidopsis thaliana, partial (82%)
Length = 1613
Score = 87.8 bits (216), Expect = 8e-18
Identities = 85/321 (26%), Positives = 136/321 (41%), Gaps = 35/321 (10%)
Frame = +3
Query: 65 VTTFGAVGDGIADDTVSFKMAWDTACQ-SELPGNVILVPKG------FSFIIQSTIFTGP 117
VT FGA GDG +T +F+ A D Q S G+ + VP G F+ T+F
Sbjct: 198 VTEFGAAGDGNTLNTKAFQSAIDHLSQYSSNGGSQLYVPPGRWLTGSFNLTSHFTLF--- 368
Query: 118 CQGGIVFKVEGTLMPPDGPESWP-------------KNISRHQWLVF-YRINGMSLEGS- 162
+ ++ WP R+ L+F + + + G+
Sbjct: 369 ------LHKDAVILGSQDESEWPVIDPLPSYGRGRDTQGGRYSSLIFGTNLTDVVITGNN 530
Query: 163 GLIDGRGEKWWDLPCKPHRQGPHWTTLPGPCDSPVAIRFFMSSNLTVQGLRVKNSPQFNF 222
G +DG+GE WW K H+ +T P I S N+ + L + NSP +N
Sbjct: 531 GTLDGQGELWWQ---KFHKGKLTYTR-------PYLIEIMYSDNIQISNLTLVNSPSWNV 680
Query: 223 RFDGCIKVHVESIYITAPKLSPNTDGIHIENTNDVKIYNSMISNGDDCVSIGSG------ 276
+ V+ I I AP SPNTDGI+ ++ + +I + I +GDDCV++ SG
Sbjct: 681 HPVYSSNIIVQGITILAPVNSPNTDGINPDSCTNTRIEDCYIVSGDDCVAVKSGWDEYGI 860
Query: 277 -----CYDVDIKNITC-GPGHG-ISIGSLGIHNSRACVSNITVRDSMMKGTDNGVRIKTW 329
+ I+ +TC P I++GS + ++ D + +++GVRIKT
Sbjct: 861 AYGMPTKQLVIRRLTCISPTSAVIALGS----EMSGGIQDVRAEDIVAINSESGVRIKTA 1028
Query: 330 QGGSGSVSGVKFSNIRMVNVR 350
G G V + + M ++
Sbjct: 1029VGRGGYVKDIYVRRMTMKTMK 1091
>TC86720 similar to PIR|T47941|T47941 hypothetical protein F2A19.90 -
Arabidopsis thaliana, partial (85%)
Length = 1719
Score = 85.5 bits (210), Expect = 4e-17
Identities = 86/319 (26%), Positives = 130/319 (39%), Gaps = 37/319 (11%)
Frame = +2
Query: 65 VTTFGAVGDGIADDTVSFKMAWDTACQ-SELPGNVILVPKG------FSFIIQSTIFTGP 117
+T FG VGDG +T +F+ A Q G+ + VP G FS T++
Sbjct: 278 LTDFGGVGDGNTSNTKAFQSAISHLSQYGSQGGSQLYVPAGKWLTGSFSLTSHFTLYLDR 457
Query: 118 CQGGIVFKVEGTLMPPDGPESWP-------------KNISRHQWLVFYRINGMSLE---- 160
+ L+ WP R + L+F G +L
Sbjct: 458 ---------DAVLLASQDITEWPVLEPLPSYGRGRDAPAGRFRSLIF----GTNLTDVIV 598
Query: 161 --GSGLIDGRGEKWWDLPCKPHRQGPHWTTLPGPCDSPVAIRFFMSSNLTVQGLRVKNSP 218
G+G IDG+G WW + HR+ +T P I S ++ + L + NSP
Sbjct: 599 TGGNGTIDGQGAFWWQ---QFHRKKLKYTR-------PYLIELMFSDSIQISNLTLLNSP 748
Query: 219 QFNFRFDGCIKVHVESIYITAPKLSPNTDGIHIENTNDVKIYNSMISNGDDCVSIGSGCY 278
+N + ++ I I AP SPNTDGI+ ++ + KI + I +GDDCV++ SG
Sbjct: 749 SWNVHPVYSSNIIIQGITIIAPISSPNTDGINPDSCTNTKIEDCYIVSGDDCVAVKSGWD 928
Query: 279 DVDIK-----------NITCGPGHGISIGSLGIHNSRACVSNITVRDSMMKGTDNGVRIK 327
+ IK +TC +I +LG S + ++ D T++GVRIK
Sbjct: 929 EYGIKFGWPTKQLVIRRLTCISPFSATI-ALGSEMSGG-IQDVRAEDITAIRTESGVRIK 1102
Query: 328 TWQGGSGSVSGVKFSNIRM 346
T G G V + M
Sbjct: 1103TAVGRGGYVKDIYVKRFTM 1159
>TC77765 similar to PIR|T47941|T47941 hypothetical protein F2A19.90 -
Arabidopsis thaliana, partial (61%)
Length = 1027
Score = 82.8 bits (203), Expect = 3e-16
Identities = 79/294 (26%), Positives = 124/294 (41%), Gaps = 33/294 (11%)
Frame = +3
Query: 68 FGAVGDGIADDTVSFKMAWDTACQSELPG-NVILVPKG------FSFIIQSTIFTGPCQG 120
FG VGDG +T +FK A Q+ G + + VP G FS I T++
Sbjct: 201 FGGVGDGKTSNTKAFKSAISHLSQNASEGGSQLYVPAGKWLTGSFSLISHFTLY------ 362
Query: 121 GIVFKVEGTLMPPDGPESWP-------------KNISRHQWLVF-YRINGMSLEG-SGLI 165
+ L+ WP RH L+F + + + G +G I
Sbjct: 363 ---LHKDAVLLASQDINEWPVIKPLPSYGRGRDAAAGRHTSLIFGTNLTDVIVTGDNGTI 533
Query: 166 DGRGEKWWDLPCKPHRQGPHWTTLPGPCDSPVAIRFFMSSNLTVQGLRVKNSPQFNFRFD 225
DG+G WW + H + +T P I S N+ + L + +SP +N
Sbjct: 534 DGQGSFWWQ---QFHNKKLKYTR-------PYLIELMFSDNIQISNLTLLDSPSWNIHPV 683
Query: 226 GCIKVHVESIYITAPKLSPNTDGIHIENTNDVKIYNSMISNGDDCVSIGSGCYDVDIK-- 283
+ ++ I I AP SPNTDGI+ ++ + KI + I +GDDCV++ SG + IK
Sbjct: 684 YSSNIIIKGITIIAPIRSPNTDGINPDSCTNTKIEDCYIVSGDDCVAVKSGWDEYGIKFG 863
Query: 284 ---------NITCGPGHGISIGSLGIHNSRACVSNITVRDSMMKGTDNGVRIKT 328
+TC + +I +LG S + ++ D T++ +RIKT
Sbjct: 864 WPTKQLVIRRLTCISPYSATI-ALGSEMSGG-IQDVRAEDITAVHTESXIRIKT 1019
>TC89269 similar to PIR|T05614|T05614 hypothetical protein F9D16.290 -
Arabidopsis thaliana, partial (60%)
Length = 923
Score = 79.0 bits (193), Expect = 4e-15
Identities = 73/267 (27%), Positives = 117/267 (43%), Gaps = 34/267 (12%)
Frame = +1
Query: 65 VTTFGAVGDGIADDTVSFKMA-WDTACQSELPGNVILVPKG------FSFIIQSTIFTGP 117
+T FG VGD ++ +F+ A + G ++ VP G F+ T++
Sbjct: 166 ITDFGGVGDSKTLNSKAFRAAIYRIQHLRRRGGTLLYVPPGVYLTDSFNLTSHMTLYLA- 342
Query: 118 CQGGIVFKVEGTLMPPDGPESWP-----------KNISRHQWLVFYRINGMS---LEG-S 162
G V K L +WP + + +++ F +G+ + G +
Sbjct: 343 --AGAVIKATQRL------RNWPLIAPLPSYGRGRELPGGRYISFIHGDGVRDVIITGEN 498
Query: 163 GLIDGRGEKWWDLPCKPHRQGPHWTTLPGPCDSPVAIRFFMSSNLTVQGLRVKNSPQFNF 222
G IDG+G+ WW++ W P + F S N+ + + KNSP +N
Sbjct: 499 GTIDGQGDVWWNM----------WRQRTLQFTRPNLVEFLNSRNIIISNVIFKNSPFWNI 648
Query: 223 RFDGCIKVHVESIYITAPKLSPNTDGIHIENTNDVKIYNSMISNGDDCVSIGSG------ 276
C V V + I AP+ SPNTDGI +++++V I +S IS GDD V++ SG
Sbjct: 649 HPVYCSNVVVRYVTILAPRDSPNTDGIDPDSSSNVCIEDSYISTGDDLVAVKSGWDAYGI 828
Query: 277 -----CYDVDIKNIT-CGPGHGISIGS 297
D+ I+ IT P GI++GS
Sbjct: 829 SYGRPSNDITIRRITGSSPFAGIALGS 909
>TC87011 weakly similar to GP|22136814|gb|AAM91751.1 unknown protein
{Arabidopsis thaliana}, partial (79%)
Length = 1405
Score = 76.6 bits (187), Expect = 2e-14
Identities = 85/342 (24%), Positives = 138/342 (39%), Gaps = 39/342 (11%)
Frame = +3
Query: 51 PPPHNCYNATGIFDVTTFGAVGDGIADDTVSFKMA-WDTACQSELPGNVILVPKG----- 104
P PH+ + FGAVGDG +T++F+ A + ++ G + VP G
Sbjct: 297 PRPHSV-------SILEFGAVGDGKTLNTIAFQNAIFYLKSFADKGGAQLYVPPGNWLTG 455
Query: 105 -FSFIIQSTIFT----------GPCQGGIVFKVEGTLMPPDGPESWPKNISRHQWLVFYR 153
F+ T+F P I+ + D P+ K++
Sbjct: 456 SFNLTSHLTLFLEKGAVIIGSQDPSHWNIIEALPSYGRGKDVPDGRYKSL---------- 605
Query: 154 INGMSLE------GSGLIDGRGEKWWDLPCKPH---RQGPHWTTLPGPCDSPVAIRFFMS 204
ING LE +G IDG G WWD K H PH + ++ F +
Sbjct: 606 INGNQLEDVVITGNNGTIDGNGMVWWD-SYKSHSLNHSRPHLGEI-------ISSDFVVV 761
Query: 205 SNLTVQGLRVKNSPQFNFRFDGCIKVHVESIYITAPKLSPNTDGIHIENTNDVKIYNSMI 264
SNLT N+P ++ C VH+++I I+ P SP T GI +++N+V I + +
Sbjct: 762 SNLTFW-----NAPAYSIHPGYCSNVHIQNISISTPPESPYTAGIVPDSSNNVCIEDCFV 926
Query: 265 SNGDDCVSIGSGCYDVDIK-------------NITCGPGHGISIGSLGIHNSRACVSNIT 311
S G D +S+ SG I ++ G IS G+ +S +
Sbjct: 927 SIGFDAISLKSGWDQYGINYGRPSEKVHIRRVHLRAFTGSAISFGT----EMSGGISKVL 1094
Query: 312 VRDSMMKGTDNGVRIKTWQGGSGSVSGVKFSNIRMVNVRNPI 353
+ + +++G+ +T +G G + + S+I M NV I
Sbjct: 1095IEHVNIFNSNSGIEFRTTKGRGGYIKEIALSHIEMENVHTAI 1220
>BI309956 weakly similar to GP|7381227|gb|A polygalacturonase {Lycopersicon
esculentum}, partial (21%)
Length = 512
Score = 76.6 bits (187), Expect = 2e-14
Identities = 44/95 (46%), Positives = 55/95 (57%), Gaps = 1/95 (1%)
Frame = +3
Query: 348 NVRNPIIIDQFYCITEG-CTNKTSAVFVSNILYTNIKGTYDVRSLPMRFACSDSIPCTHL 406
NV NPIIIDQ+YC + C N+TSAV V NI + NI+GT ++FACSD+ PC L
Sbjct: 12 NVSNPIIIDQYYCDSRHPCKNQTSAVQVGNISFINIQGT-SATEETIKFACSDASPCEGL 188
Query: 407 ALSDIELLPAEGDIAHDPFCWNAYGDLQTLTIPPV 441
L +I L G +CW A+G Q PPV
Sbjct: 189 YLENIFLRSYFGGNTRS-YCWQAHGSAQGYVYPPV 290
>TC82168 weakly similar to GP|11762132|gb|AAG40344.1 AT3g62110 {Arabidopsis
thaliana}, partial (33%)
Length = 587
Score = 74.7 bits (182), Expect = 7e-14
Identities = 56/163 (34%), Positives = 80/163 (48%), Gaps = 13/163 (7%)
Frame = +3
Query: 204 SSNLTVQGLRVKNSPQFNFRFDGCIKVHVESIYITAPKLSPNTDGIHIENTNDVKIYNSM 263
S N+ V L +NSP + C V ++ + I AP +PNTDGI +++ +V I ++
Sbjct: 48 SENVLVSNLTFRNSPFWTIHPVYCSNVVIKGMTILAPLNAPNTDGIDPDSSTNVCIEDNY 227
Query: 264 ISNGDDCVSIGSGCYDVDI------KNI-------TCGPGHGISIGSLGIHNSRACVSNI 310
I +GDD V+I SG I NI T G+ IGS +SNI
Sbjct: 228 IESGDDLVAIKSGWDQYGIAVAKPSTNIIVSRVSRTTPTCSGVGIGS----EMSGGISNI 395
Query: 311 TVRDSMMKGTDNGVRIKTWQGGSGSVSGVKFSNIRMVNVRNPI 353
T+ + + + GVRIK+ G G + V SNIRM V+ PI
Sbjct: 396 TIENLHVWNSAAGVRIKSDNGRGGYIKNVSISNIRMERVKIPI 524
>TC79890 similar to GP|11762132|gb|AAG40344.1 AT3g62110 {Arabidopsis
thaliana}, partial (55%)
Length = 1151
Score = 71.2 bits (173), Expect = 8e-13
Identities = 60/200 (30%), Positives = 89/200 (44%), Gaps = 12/200 (6%)
Frame = +3
Query: 162 SGLIDGRGEKWWDLPCKPHRQGPHWTTLPGPCDSPVAIRFFMSSNLTVQGLRVKNSPQFN 221
+G IDG+G WW K + T P + S+ + + NSP +
Sbjct: 57 NGTIDGQGSIWWS---KFRNKTLDHTR-------PHLVELINSTEVLISNATFLNSPFWT 206
Query: 222 FRFDGCIKVHVESIYITAPKLSPNTDGIHIENTNDVKIYNSMISNGDDCVSIGSGCYDVD 281
C V V+++ I P SPNTDGI +++++V I + IS GDD +SI SG +
Sbjct: 207 IHPVYCSNVTVQNVTIIVPFGSPNTDGIDPDSSDNVCIEDCYISTGDDLISIKSGWDEYG 386
Query: 282 IK------NITC------GPGHGISIGSLGIHNSRACVSNITVRDSMMKGTDNGVRIKTW 329
I NI+ GI+IGS VS + D + + + +RIKT
Sbjct: 387 ISFGRPSTNISIHRLTGRTTSAGIAIGS----EMSGGVSEVYAEDIYIFDSKSAIRIKTS 554
Query: 330 QGGSGSVSGVKFSNIRMVNV 349
G G V V SN+ ++NV
Sbjct: 555 PGRGGYVRNVYISNMTLINV 614
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.138 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,569,301
Number of Sequences: 36976
Number of extensions: 330854
Number of successful extensions: 3406
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 2930
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3248
length of query: 460
length of database: 9,014,727
effective HSP length: 99
effective length of query: 361
effective length of database: 5,354,103
effective search space: 1932831183
effective search space used: 1932831183
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0122.9


