

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0122.3
(1172 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
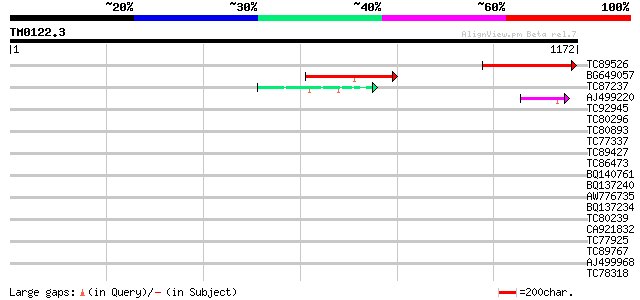
Score E
Sequences producing significant alignments: (bits) Value
TC89526 similar to GP|14596139|gb|AAK68797.1 Unknown protein {Ar... 320 2e-87
BG649057 similar to PIR|H86201|H86 hypothetical protein [importe... 288 1e-77
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 45 2e-04
AJ499220 similar to SP|Q09863|YAF Hypothetical protein C29E6.10c... 44 4e-04
TC92945 similar to GP|22137072|gb|AAM91381.1 At1g42440/F7F22_7 {... 40 0.007
TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported]... 38 0.020
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 38 0.026
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 37 0.058
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 36 0.076
TC86473 similar to PIR|E84828|E84828 probable WD-40 repeat prote... 36 0.076
BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus... 36 0.099
BQ137240 similar to PIR|F75311|F753 ABC transporter ATP-binding... 36 0.099
AW776735 homologue to PIR|T19148|T19 hypothetical protein C09G5.... 36 0.099
BQ137234 similar to EGAD|35719|3709 hypothetical protein {Burkho... 35 0.13
TC80239 similar to GP|7208779|emb|CAB76912.1 hypothetical protei... 35 0.13
CA921832 similar to GP|14194103|gb AT3g22390/MCB17_12 {Arabidops... 35 0.17
TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1 ... 35 0.17
TC89767 similar to GP|9759165|dbj|BAB09721.1 gene_id:MEE6.26~unk... 34 0.29
AJ499968 weakly similar to GP|23495961|gb| hypothetical protein ... 34 0.29
TC78318 similar to PIR|T45852|T45852 hypothetical protein F3A4.7... 34 0.29
>TC89526 similar to GP|14596139|gb|AAK68797.1 Unknown protein {Arabidopsis
thaliana}, partial (40%)
Length = 1105
Score = 320 bits (821), Expect = 2e-87
Identities = 156/193 (80%), Positives = 175/193 (89%)
Frame = +1
Query: 978 EGIARCTFEDKILMSDIVFLRAWTQVEVPQFYNPLTTALQPRDQTWKGMRTVAELRRDHD 1037
EGIARCTFEDKILMSDIVFLRAWTQVEVPQFYNPLTTALQPRDQTWKGMRTVAELRR+H+
Sbjct: 1 EGIARCTFEDKILMSDIVFLRAWTQVEVPQFYNPLTTALQPRDQTWKGMRTVAELRREHN 180
Query: 1038 LPVPVNKDSLYKKIERKPRKFNPLVVPRSLQERLPFASKPKETPKRNQPLLEQRRQKGVV 1097
LP+PVNKDSLYKKIERKPRKFNPLV+P+SLQ LPF SKPK TPKR + + RRQKGVV
Sbjct: 181 LPIPVNKDSLYKKIERKPRKFNPLVIPKSLQANLPFESKPKHTPKRKRLSFDDRRQKGVV 360
Query: 1098 MEPRERKVLALVQHIQLINHDKMKKRKLKENEKRKAHEAEKAKEEQLSKKRRREERREKY 1157
+EPRERK+ ALVQH+QL+ +K+KKRK KE EKRK EAE+AKEE +SKKRRREERR+KY
Sbjct: 361 VEPRERKIHALVQHLQLMKTEKIKKRKHKEGEKRKVLEAERAKEELVSKKRRREERRDKY 540
Query: 1158 RTQDKSSKKMRRS 1170
RTQDK +KK+RR+
Sbjct: 541 RTQDKLNKKIRRA 579
>BG649057 similar to PIR|H86201|H86 hypothetical protein [imported] -
Arabidopsis thaliana, partial (8%)
Length = 644
Score = 288 bits (736), Expect = 1e-77
Identities = 150/214 (70%), Positives = 169/214 (78%), Gaps = 23/214 (10%)
Frame = +2
Query: 612 LNDGNVNTEDYSKCAQFMDKRWDKKDNEEIRNRFVTGNLAKAALRNGLPKATTEEE---- 667
L+DG V+TED SKCA+ M ++WD+KD+ EIRNRFV+GNLAKAA RN L KA TEEE
Sbjct: 2 LDDGMVHTEDCSKCAKLMSQKWDEKDHGEIRNRFVSGNLAKAARRNALQKANTEEEEEDE 181
Query: 668 NDDVYGDFEDLETGEKIENNQTDDA----THNGDDLEAETRRLKKLAL------------ 711
++DVYGDFEDLETGE EN +TDDA T G D EAE RRLKKLAL
Sbjct: 182 DEDVYGDFEDLETGENHENYKTDDAFAITTQKGVDREAEERRLKKLALHAKFVSRYDDDP 361
Query: 712 ---QEDGGNENEAKFRRGQPNETSYFDKLKEEIELQKQMNIAELNDLDEATRLEIEGFRT 768
+ED GNENEAKF R QPNE++Y DKLKEEIEL+KQMNIAELNDLDE TRLE+EGFRT
Sbjct: 362 ETPEEDTGNENEAKFHREQPNESNYIDKLKEEIELRKQMNIAELNDLDEDTRLEVEGFRT 541
Query: 769 GTYVRLEVHDVPCEMVEYFDPYHPILIGGVGLGE 802
GTY+RLEVHDVPCEMVE+FDPYHPIL+GGVGLGE
Sbjct: 542 GTYLRLEVHDVPCEMVEHFDPYHPILVGGVGLGE 643
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 44.7 bits (104), Expect = 2e-04
Identities = 63/270 (23%), Positives = 106/270 (38%), Gaps = 23/270 (8%)
Frame = +1
Query: 513 DSSGNEDGSASDDGASSDSESLNEEEEDDVMGNVSKWKESLAERTLSQKTPSLMQLVYGE 572
+ S E+ + + G SS+ +S E EE G S+ K L + S + E
Sbjct: 4 EKSHLENEESKETGESSEEKSHLENEESKETGESSEEKSHLENEENKDEEKSKQE---NE 174
Query: 573 STVNLISINKENDSSEDEESDGDFFEPIEEVKKKNVRDGL--NDGNVN-----TEDYSKC 625
+ I +EN+ ++DEE E ++ +K + L N+G TE+ SK
Sbjct: 175 EIKDGEKIQQENEENKDEEKSQQENEENKDEEKSQQENELKKNEGGEKETGEITEEKSKQ 354
Query: 626 AQFMDKRWDKKDNEEIRNRFVTGNLAKAALRNGL---PKATTEEENDDVYGDFED----- 677
+ KD E + G+ AK + K TEE N G+ E+
Sbjct: 355 ENEETSETNSKDKENEESN-QNGSDAKEQVGENHEQDSKQGTEETNGTEGGEKEEHDKIK 531
Query: 678 --------LETGEKIENNQTDDATHNGDDLEAETRRLKKLALQEDGGNENEAKFRRGQPN 729
++ GEK NN+ + ++GD+ + K ++ N+ E +F +
Sbjct: 532 EDTSSDNQVQDGEK--NNEAREENYSGDNASSAVVDNK----SQESSNKTEEQFDK---- 681
Query: 730 ETSYFDKLKEEIELQKQMNIAELNDLDEAT 759
K K E EL+ Q N E + ++T
Sbjct: 682 ------KEKNEFELESQKNSNETTESTDST 753
Score = 34.3 bits (77), Expect = 0.29
Identities = 75/452 (16%), Positives = 160/452 (34%), Gaps = 55/452 (12%)
Frame = +1
Query: 369 KVDDENSDITRKGNERDIGEVLVRSLQETKYPINEKLENSMINLFAQSSEALAEAQGANM 428
K EN + + ++ E ++ + K + E E Q +E +G
Sbjct: 343 KSKQENEETSETNSKDKENEESNQNGSDAKEQVGENHEQDS----KQGTEETNGTEGGEK 510
Query: 429 DVEQDGIIETVNYNEMDSDG--GSESSDQDEADAMTGSGLPDQAEDDAAGDKSTNKDHLE 486
+ E D I E + + DG +E+ +++ + S + D +++ D E
Sbjct: 511 E-EHDKIKEDTSSDNQVQDGEKNNEAREENYSGDNASSAVVDNKSQESSNKTEEQFDKKE 687
Query: 487 EHIEFHNGRRRRRAIFGNDADQSDLMDSSGNED-------------GSASDDGASSDSES 533
++ EF ++ D + +S GNE GSAS+ +
Sbjct: 688 KN-EFELESQKNSNETTESTDSTITQNSQGNESEKDQAQTENDTPKGSASESDEQKQEQE 864
Query: 534 LNEEEEDDVM--------GNVSKWKESLAERTLSQK---------TPSLMQLVYGES--T 574
N +DDV GN + K++ + K TP+ G +
Sbjct: 865 QNNTTKDDVQTTDTSSQNGNDTTEKQNETSEDANSKKEDSSALNTTPNNEDSKSGVAGDQ 1044
Query: 575 VNLISINKENDSSEDEESDGDFFEPIEEVKKKNV-RDGLND--GNVNTEDYSKCAQ---- 627
+ + +++ + + G++ + E +KN ++G + + NT D A
Sbjct: 1045ADSTTTTSSSETQDGNTNHGEYKDTTNENPEKNSGQEGTQESGSSSNTFDNKDAASNKVQ 1224
Query: 628 -------FMDKRWDKKDNEEIRNRFVTGNLAKAALRNGLPKATTEEENDDVYGDFEDLET 680
+++ D+ + E ++ + A + N TT +E+ + D + T
Sbjct: 1225LTTTSDTSSEQKKDESSSAESKSESSQNDNANSGQSN-----TTSDESANDNKDSSQVTT 1389
Query: 681 GEKIENNQTDDATHNGDDLEAETRRLKKLALQEDGGNENEAKFRRGQPNETSYF------ 734
+ + +N D+ + +++ + + G N+A Q +
Sbjct: 1390SSENSAEGNSNTENNSDENQNDSKNNEN--TNDSGNTSNDANVNENQNENAAQTKTSENE 1563
Query: 735 -DKLKEEIELQKQMNIAELNDLDEATRLEIEG 765
D E +E +K+ N + D+D + +G
Sbjct: 1564GDAQNESVESKKENNESAHKDVDNNSNSNDQG 1659
>AJ499220 similar to SP|Q09863|YAF Hypothetical protein C29E6.10c in chromosome
I. [Fission yeast] {Schizosaccharomyces pombe}, partial
(12%)
Length = 518
Score = 43.9 bits (102), Expect = 4e-04
Identities = 32/109 (29%), Positives = 55/109 (50%), Gaps = 8/109 (7%)
Frame = -3
Query: 1056 RKFNPLVVPRSLQERLPFASKPKETPKRNQPLLEQRRQKGVVMEPRERKVLALVQHIQLI 1115
R+ + R ++R+ A + K +R Q LLE+ ++ + E RE K L + +
Sbjct: 357 RRMFQIFAARMFEQRVLNAYREKVAQERQQKLLEELEEENRLKEERELKKLKEKERKKAK 178
Query: 1116 NHDKMKKRKLKENEKR--------KAHEAEKAKEEQLSKKRRREERREK 1156
N +K++K +E +R KA AE+ K+ + +KRR EER +K
Sbjct: 177 NR-ALKQQKEEERARREAERLAEEKAQRAEREKKLEEERKRREEERLKK 34
>TC92945 similar to GP|22137072|gb|AAM91381.1 At1g42440/F7F22_7 {Arabidopsis
thaliana}, partial (13%)
Length = 763
Score = 39.7 bits (91), Expect = 0.007
Identities = 16/51 (31%), Positives = 33/51 (64%)
Frame = +2
Query: 909 IVKKIKLVGYPCKIFKKTALIKEMFTSDLEIARFEGAAIRTVSGIRGQVKK 959
I+K++ L GYP ++ K+ A ++ MF + ++ F+ + T G+RG++K+
Sbjct: 182 ILKRVILTGYPQRVSKRKASVRHMFYNPEDVKWFKPVELYTKRGLRGRIKE 334
>TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported] -
Arabidopsis thaliana, partial (34%)
Length = 1734
Score = 38.1 bits (87), Expect = 0.020
Identities = 42/187 (22%), Positives = 81/187 (42%), Gaps = 8/187 (4%)
Frame = +3
Query: 429 DVEQDGIIETVNYNEMDSDGGSESSDQDEADAMTGSGLPDQAEDDAAGDKSTNKDHLEEH 488
DVEQ+ + D GS+++ +D+ ++ D+ D D E+H
Sbjct: 762 DVEQE--------SSHSDDCGSDNAQEDDQVSLNSDDDNQLGSDNTGSDD----DEAEDH 905
Query: 489 IEFHNGRRRRRAIFGNDADQSDLMDSSGNEDGSASDDGASSDSESLNEEEEDDVMGNVSK 548
+ R + + D +D ++ G++ + +DG S+ E + + ++G+V
Sbjct: 906 DGVSDDENDRSSDYETSGDDADNVEDEGDDLEDSEEDGGISEHEG---DGDLHILGSVDT 1076
Query: 549 WKESLAERTLSQKTPSLM-QLVYGESTV----NLISINKENDSSEDEE---SDGDFFEPI 600
K +L + +K QL +S++ L EN E+E+ S+ D F+ I
Sbjct: 1077-KTTLKDLAKKRKFSDFNDQLTAADSSLRALKKLAGTTMENALPENEDGILSNAD-FQRI 1250
Query: 601 EEVKKKN 607
+E+K KN
Sbjct: 1251KELKAKN 1271
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 37.7 bits (86), Expect = 0.026
Identities = 26/101 (25%), Positives = 39/101 (37%)
Frame = +3
Query: 441 YNEMDSDGGSESSDQDEADAMTGSGLPDQAEDDAAGDKSTNKDHLEEHIEFHNGRRRRRA 500
Y E SD + D D+ D Q EDD ++ + D EE
Sbjct: 300 YKENKSDTEDDEDDDDDDDV--------QDEDDDGEEEDYSGDEGEEE------------ 419
Query: 501 IFGNDADQSDLMDSSGNEDGSASDDGASSDSESLNEEEEDD 541
G+ D + + G++DG DD + E E+EED+
Sbjct: 420 --GDPEDDPEANGAGGSDDGEDDDDDGDEEDEEDGEDEEDE 536
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 36.6 bits (83), Expect = 0.058
Identities = 30/111 (27%), Positives = 45/111 (40%), Gaps = 14/111 (12%)
Frame = +3
Query: 445 DSDGGSESSDQDEADAMTGSGLPDQAEDDAA-------GDKSTNKDHLEEHIEFHNGRRR 497
D++G + D+DE D G G + ED+ + G+ S NK + ++ E G
Sbjct: 408 DAEGEDGNDDEDEEDD-DGDGAFGEGEDELSSEDGGGYGNNSNNKSNSKKAPEGGAGGAD 584
Query: 498 RRAIFGNDADQSDL-------MDSSGNEDGSASDDGASSDSESLNEEEEDD 541
+D D D D E+G D+ D E EEEED+
Sbjct: 585 ENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEEEEEDE 737
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 36.2 bits (82), Expect = 0.076
Identities = 32/109 (29%), Positives = 46/109 (41%)
Frame = +1
Query: 443 EMDSDGGSESSDQDEADAMTGSGLPDQAEDDAAGDKSTNKDHLEEHIEFHNGRRRRRAIF 502
E + DG D DE D + D ++D +GD+ ++D E NG
Sbjct: 283 EENKDGSETEDDDDEDDDDDVNDEDDDNDEDFSGDED-DEDADPEDDPVPNG-------- 435
Query: 503 GNDADQSDLMDSSGNEDGSASDDGASSDSESLNEEEEDDVMGNVSKWKE 551
A SD D ++D +DDG D + EEE+DD SK K+
Sbjct: 436 ---AGGSDDDDEDDDDDDDDNDDGEDEDED---EEEDDDEDQPPSKKKK 564
>TC86473 similar to PIR|E84828|E84828 probable WD-40 repeat protein
[imported] - Arabidopsis thaliana, partial (68%)
Length = 2654
Score = 36.2 bits (82), Expect = 0.076
Identities = 46/180 (25%), Positives = 69/180 (37%), Gaps = 7/180 (3%)
Frame = +2
Query: 445 DSDGGSESSDQDEADAMTGSGLPDQAEDDAAGDKSTNKDHLEEHIEFHNGRRRRRAIFGN 504
DSD E SD D S +DD++ S +K LE+ E + A +
Sbjct: 140 DSDDSVEVSDYDSISEDEDSSADSIDDDDSSDSDSDSKSELEDE-EGASPTTHHNASDDS 316
Query: 505 DA--DQSDLMDSSGNEDGS---ASDDGASSDSESLNEEEEDDVMGNVS-KWKESLAERTL 558
D+ ++ D++DS G + S GA SDS S +E + +G V KW E
Sbjct: 317 DSQDEEEDVVDSEGGSESSDLHQEGGGAESDS-SEDEVAPRNTIGEVPLKWYED------ 475
Query: 559 SQKTPSLMQLVYGESTVNLISINKENDSSEDEESDGDFFEPIEEVKK-KNVRDGLNDGNV 617
E + K+ E E+ G F +++ K + V D ND V
Sbjct: 476 -------------EPHIGYDIKGKKIKKKEREDKLGSFLANVDDSKNWRKVFDEYNDEEV 616
>BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus
norvegicus}, partial (9%)
Length = 620
Score = 35.8 bits (81), Expect = 0.099
Identities = 34/114 (29%), Positives = 49/114 (42%), Gaps = 17/114 (14%)
Frame = +1
Query: 444 MDSDG-GSESSDQDEADAMTGSGLPDQAEDDAAGDKSTNKDHL---EEHIEFHNGRRRRR 499
MD D S+SSD D +D+ GS A DD+ ++ D EE + + ++
Sbjct: 226 MDVDEKSSDSSDSDSSDSSDGSDSDSDASDDSDDSSDSSSDDSSSDEEPVPAPKKEKAKK 405
Query: 500 AIFGND-ADQSDLMDSSGNEDGS----------ASDD--GASSDSESLNEEEED 540
A + SD D S +E S SDD ASSDS+S ++ D
Sbjct: 406 AKKAKSVSSSSDSSDDSSSESESEPEPXNVPLPESDDSSSASSDSDSSSDSSSD 567
Score = 32.7 bits (73), Expect = 0.84
Identities = 27/96 (28%), Positives = 42/96 (43%), Gaps = 6/96 (6%)
Frame = +1
Query: 506 ADQSDLMD------SSGNEDGSASDDGASSDSESLNEEEEDDVMGNVSKWKESLAERTLS 559
A QSD MD S + D S S DG+ SDS++ ++ DD + S S E +
Sbjct: 208 AAQSDAMDVDEKSSDSSDSDSSDSSDGSDSDSDA--SDDSDDSSDSSSDDSSSDEEPVPA 381
Query: 560 QKTPSLMQLVYGESTVNLISINKENDSSEDEESDGD 595
K + S++ +DSS+D S+ +
Sbjct: 382 PKKEK------AKKAKKAKSVSSSSDSSDDSSSESE 471
Score = 32.3 bits (72), Expect = 1.1
Identities = 22/90 (24%), Positives = 40/90 (44%)
Frame = +1
Query: 504 NDADQSDLMDSSGNEDGSASDDGASSDSESLNEEEEDDVMGNVSKWKESLAERTLSQKTP 563
+D+D SD D S ++ ++ D SSDS S + +++ + K K A++ S +
Sbjct: 256 SDSDSSDSSDGSDSDSDASDDSDDSSDSSSDDSSSDEEPVPAPKKEKAKKAKKAKSVSSS 435
Query: 564 SLMQLVYGESTVNLISINKENDSSEDEESD 593
S + +DSS + ES+
Sbjct: 436 S----------------DSSDDSSSESESE 477
>BQ137240 similar to PIR|F75311|F753 ABC transporter ATP-binding protein -
Deinococcus radiodurans (strain R1), partial (4%)
Length = 1094
Score = 35.8 bits (81), Expect = 0.099
Identities = 19/89 (21%), Positives = 47/89 (52%)
Frame = +1
Query: 1078 KETPKRNQPLLEQRRQKGVVMEPRERKVLALVQHIQLINHDKMKKRKLKENEKRKAHEAE 1137
K T Q + QR++KG + E+K + Q NH++ KK ++ + +R+ +
Sbjct: 61 KRTKNTKQLTIRQRKEKGWNPDGTEKKTIHKQQKTTRKNHER-KKEPVRGSRERREARSV 237
Query: 1138 KAKEEQLSKKRRREERREKYRTQDKSSKK 1166
KE++ ++R+++ ++++ T + ++
Sbjct: 238 GGKEKKGVERRKKKTLKQEHETDKRIERE 324
>AW776735 homologue to PIR|T19148|T19 hypothetical protein C09G5.8 -
Caenorhabditis elegans, partial (1%)
Length = 467
Score = 35.8 bits (81), Expect = 0.099
Identities = 22/69 (31%), Positives = 37/69 (52%)
Frame = +2
Query: 9 SNKAHRIRQSGAKSFKKKQNKKKKQDDDDGLIDKPQNPRAFAFSSTNKAKRLKSRAVEKE 68
SNK HR++ + A+ ++ +++K DDD L+D+ Q+ R + R +S AV K
Sbjct: 146 SNKDHRVKSTSAEQEQQSAVQRRKGDDD--LLDELQDNRIHTVARRRDVSRSRSPAVNK- 316
Query: 69 QRRLHVPIT 77
+H IT
Sbjct: 317 ---MHFTIT 334
>BQ137234 similar to EGAD|35719|3709 hypothetical protein {Burkholderia
cepacia}, partial (6%)
Length = 1184
Score = 35.4 bits (80), Expect = 0.13
Identities = 23/73 (31%), Positives = 36/73 (48%)
Frame = +1
Query: 1090 QRRQKGVVMEPRERKVLALVQHIQLINHDKMKKRKLKENEKRKAHEAEKAKEEQLSKKRR 1149
+R+ +G E +++K I DK KKR+ +E EK + E+ K + K+RR
Sbjct: 955 KRQARGARDEKKQQKETTERAQIS----DKRKKREKRETEKDQQTTRERKKRQ*RRKQRR 1122
Query: 1150 REERREKYRTQDK 1162
R R K R Q +
Sbjct: 1123 RAAERRKRRRQQR 1161
>TC80239 similar to GP|7208779|emb|CAB76912.1 hypothetical protein {Cicer
arietinum}, partial (43%)
Length = 930
Score = 35.4 bits (80), Expect = 0.13
Identities = 19/55 (34%), Positives = 31/55 (55%), Gaps = 2/55 (3%)
Frame = +3
Query: 1119 KMKKRKLKENEKRKAHEAEKAKEEQLSKKRRRE--ERREKYRTQDKSSKKMRRSE 1171
+ K+ + K EK++ E EKAKE KKR E ++R Y+ Q ++ +K + E
Sbjct: 123 RKKEEEAKLKEKKRLEEIEKAKEALQRKKRNAEKAQQRALYKAQKEAEQKEKERE 287
>CA921832 similar to GP|14194103|gb AT3g22390/MCB17_12 {Arabidopsis
thaliana}, partial (3%)
Length = 802
Score = 35.0 bits (79), Expect = 0.17
Identities = 26/97 (26%), Positives = 40/97 (40%)
Frame = -1
Query: 445 DSDGGSESSDQDEADAMTGSGLPDQAEDDAAGDKSTNKDHLEEHIEFHNGRRRRRAIFGN 504
D DGG E + P + D +G K +++ E GR +RR
Sbjct: 514 DDDGGMEMQE------------PTRLRDRGSGKKERDRERERERERDRLGRNKRR----- 386
Query: 505 DADQSDLMDSSGNEDGSASDDGASSDSESLNEEEEDD 541
++D + EDG G + ES+N+EE+DD
Sbjct: 385 ---RNDRLMHGVREDG-----GEDTSEESINDEEDDD 299
>TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1
{Arabidopsis thaliana}, partial (47%)
Length = 1538
Score = 35.0 bits (79), Expect = 0.17
Identities = 41/207 (19%), Positives = 89/207 (42%), Gaps = 14/207 (6%)
Frame = +3
Query: 373 ENSDITRKGNERDIGEVL-VRSLQETKYPI-----NEKLENSMINLFAQSSEALAEAQGA 426
EN D+ G +I E++ V+ +E K +E+ + S L + ++ +++ +
Sbjct: 453 ENDDLEDYGKPEEISEIIDVKKDKEIKQKAKSESESEESKESDSELRKKRRKSYKKSRES 632
Query: 427 NMDVEQDGIIE------TVNYNEMDSDGGSESSDQDEADAMTGSGLPDQAEDDAAGDKST 480
+ + E + +E + Y+E DSD SE D+ S ++ +
Sbjct: 633 DSESESESEVEDRKRRKSRKYSESDSDTNSEEEDRKRRRRRKSSRRRRKSSRKKRRCSDS 812
Query: 481 NKDHLEEHIEF-HNGRRRRRAIFGNDA-DQSDLMDSSGNEDGSASDDGASSDSESLNEEE 538
++ ++ + +GR+R+R+ + +S S G+E+ D A + ++
Sbjct: 813 DESETDDESGYDDSGRKRKRSKRSRKSKKKSSEPVSEGSEEIELGSDSAVAKINEEIDDV 992
Query: 539 EDDVMGNVSKWKESLAERTLSQKTPSL 565
+++ M ++ L E SQK P+L
Sbjct: 993 DEEKMAEMNAEALKLKELFESQKKPAL 1073
>TC89767 similar to GP|9759165|dbj|BAB09721.1 gene_id:MEE6.26~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 1587
Score = 34.3 bits (77), Expect = 0.29
Identities = 51/242 (21%), Positives = 94/242 (38%), Gaps = 11/242 (4%)
Frame = +2
Query: 366 QFTKVDDENSDITRKGNERDIGEVLVRSLQETKYPINEKLENSMINLFAQSSEALAEAQG 425
++ ++ E D G+ + LQ+ I + + S + L + E L E
Sbjct: 632 EWEALEQETEDNANSGSR-------ILPLQDLSLNILPQDDQSEVGLVEHTGETLLEILE 790
Query: 426 ANMDVEQDGIIETVNY----------NEMDSDGGSESSDQDEADAMTGSGLPDQAEDDAA 475
+ E+ G + NY +M +DG S Q + D G +P A
Sbjct: 791 GD---ERGGSMRRKNYIPKKKVINIEGKMVADGIDAS--QGQVDDNGGDWMP-------A 934
Query: 476 GDKSTNKDHLEEHIEFHNGRRRRRAIFGNDADQSDLMDSSGNEDGSASDDGASSDSESLN 535
+ST++ L R+ RR + + D D N DGS +D +S+ +
Sbjct: 935 VSRSTHRRFLR--------RKARREHYDALSSNQDQQDMEENIDGSVCEDDKTSNLDVHQ 1090
Query: 536 EEEEDDVMGNVSKWKESLAERTLSQK-TPSLMQLVYGESTVNLISINKENDSSEDEESDG 594
++E + VSK AE + + +L Q+ E ++ +++ + S E+S
Sbjct: 1091SDDEKHIENVVSKDDMIFAENNDGETISATLKQMTLEEGSLEVLN-EEHKPSLSPEDSRA 1267
Query: 595 DF 596
+F
Sbjct: 1268NF 1273
>AJ499968 weakly similar to GP|23495961|gb| hypothetical protein {Plasmodium
falciparum 3D7}, partial (19%)
Length = 342
Score = 34.3 bits (77), Expect = 0.29
Identities = 24/75 (32%), Positives = 42/75 (56%), Gaps = 1/75 (1%)
Frame = -3
Query: 1099 EPRERKVLALVQHIQLINHDKMKKRKLKENEKRKAHEAEK-AKEEQLSKKRRREERREKY 1157
E RERK + + I +K ++ + KE E+R+ E E+ AKE++ +++ +EER E+
Sbjct: 334 EERERKEREEREEQERIAKEKEEQER-KEREERERLELERIAKEKEERERKEKEEREERE 158
Query: 1158 RTQDKSSKKMRRSEA 1172
R + + R EA
Sbjct: 157 RLEAVVRIEKERKEA 113
>TC78318 similar to PIR|T45852|T45852 hypothetical protein F3A4.70 -
Arabidopsis thaliana, partial (48%)
Length = 1923
Score = 34.3 bits (77), Expect = 0.29
Identities = 22/57 (38%), Positives = 33/57 (57%), Gaps = 2/57 (3%)
Frame = +1
Query: 1118 DKMKKRKLKENE--KRKAHEAEKAKEEQLSKKRRREERREKYRTQDKSSKKMRRSEA 1172
DK+K ++ E KRK H E +E++ K +EERRE RT+ K K++ + EA
Sbjct: 1396 DKVKDATTEKTEQYKRKQHGLESKEEKKERKAAVKEERREARRTK-KEMKELYKCEA 1563
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,243,980
Number of Sequences: 36976
Number of extensions: 399523
Number of successful extensions: 2140
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 2023
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2098
length of query: 1172
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1065
effective length of database: 5,058,295
effective search space: 5387084175
effective search space used: 5387084175
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0122.3


