

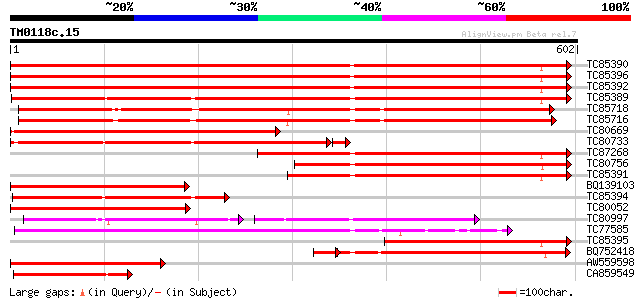
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0118c.15
(602 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 799 0.0
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 794 0.0
TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chape... 792 0.0
TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protei... 632 0.0
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 486 e-138
TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chape... 461 e-130
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 441 e-124
TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa prot... 433 e-124
TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chape... 412 e-115
TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 379 e-105
TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chape... 295 3e-80
BQ139103 homologue to GP|2655420|gb| heat shock cognate protein ... 295 4e-80
TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic retic... 285 4e-77
TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chapero... 254 9e-68
TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [im... 164 4e-65
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 239 2e-63
TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protei... 236 1e-62
BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated pr... 219 2e-61
AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 ... 211 5e-55
CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus sto... 181 7e-46
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 799 bits (2063), Expect = 0.0
Identities = 409/610 (67%), Positives = 489/610 (80%), Gaps = 14/610 (2%)
Frame = +1
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
MA + AIGIDLGTTYSCV VW H+RVEII N+ GNRTTPSYVAFTDTERL+GDAA N
Sbjct: 136 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 315
Query: 61 QLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAA 120
Q+A+NP NTVFDAKRLIGRRF D SVQ DMKLWPFKV+ G DKPMI V YK EE++ +A
Sbjct: 316 QVAMNPTNTVFDAKRLIGRRFGDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQFSA 495
Query: 121 EEISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEP 180
EEISSMVL KMKE+AEAYLG +K AV+TVPAYFN++QRQ+TKDAG I+G +V+RIINEP
Sbjct: 496 EEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVLRIINEP 675
Query: 181 TAASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFD 240
TAA+IAYGLDKK GE+NVL+FDLGGGTFDVSL+TI+EGIF+V AT GDT+LGG DFD
Sbjct: 676 TAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 855
Query: 241 NNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLH 300
N +VN V F+RK+ KDI+ N ++LRRLR ACE+AKR LSST+QTTIE+DSL GID +
Sbjct: 856 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFY 1035
Query: 301 VTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMF 360
T+T+ FE+LN DLF KCME VEKCL +AK+ K+ VH++VLVGGSTRIPKVQQLL++ F
Sbjct: 1036TTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFF 1215
Query: 361 SVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVM 420
+ KELCKSINPDEAVAYGAAVQAAILS EG++KV++LLLLDV PLSLG+E GVM
Sbjct: 1216N----GKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVM 1383
Query: 421 SVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKG 480
+VLIP+NTTIPTKKE+ F T SDNQ V I+VYEGE +T N LGKFELSG PAP+G
Sbjct: 1384TVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRG 1563
Query: 481 DTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDE 540
+I VCFD+DA+GI+ VSAED + K ITITN GRLS+EE+ +MV++A +YK+EDE
Sbjct: 1564VPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDE 1743
Query: 541 EVRKKVKAKNSLENYAYEARDRV--------------KKLEKMVEEVIEWLDRNQLAEAE 586
E +KKV+AKN+LENYAY R+ + KK+E +E I+WLD NQL EA+
Sbjct: 1744EHKKKVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEAD 1923
Query: 587 EFEYKKQELE 596
EFE K +ELE
Sbjct: 1924EFEDKMKELE 1953
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 794 bits (2051), Expect = 0.0
Identities = 407/610 (66%), Positives = 488/610 (79%), Gaps = 14/610 (2%)
Frame = +2
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
MA + AIGIDLGTTYSCV VW H+RVEII N+ GNRTTPSYVAFTD+ERL+GDAA N
Sbjct: 107 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 286
Query: 61 QLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAA 120
Q+A+NP NTVFDAKRLIGRR SD SVQ DMKLWPFKV+ G +KPMI V YKGEE+ A+
Sbjct: 287 QVAMNPTNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEEKLFAS 466
Query: 121 EEISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEP 180
EEISSMVL KM+E+AEAYLG +K AV+TVPAYFN++QRQ+TKDAG IAG +V+RIINEP
Sbjct: 467 EEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 646
Query: 181 TAASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFD 240
TAA+IAYGLDKK GE+NVL+FDLGGGTFDVSL+TI+EGIF+V AT GDT+LGG DFD
Sbjct: 647 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 826
Query: 241 NNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLH 300
N +V V F+RK+ KDI+ N ++LRRLR ACE+AKR LSST+QTTIE+DSL G+D +
Sbjct: 827 NRMVTHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVDFY 1006
Query: 301 VTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMF 360
T+T+ FE+LN DLF KCME VEKCL +AK+ KS VH++VLVGGSTRIPKVQQLL++ F
Sbjct: 1007TTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQDFF 1186
Query: 361 SVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVM 420
+ KELCKSINPDEAVAYGAAVQAAILS EG++KV++LLLLDV PLS G+E GVM
Sbjct: 1187N----GKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSQGLETAGGVM 1354
Query: 421 SVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKG 480
+VLIP+NTTIPTKKE+ F T SDNQ V I+VYEGE +T N LGKFELSG PAP+G
Sbjct: 1355TVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTKDNNLLGKFELSGIPPAPRG 1534
Query: 481 DTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDE 540
+I VCFD+DA+GI+ VSAED + K ITITN GRLS+EE+ +MV++A +YK+EDE
Sbjct: 1535VPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDE 1714
Query: 541 EVRKKVKAKNSLENYAYEARDRV--------------KKLEKMVEEVIEWLDRNQLAEAE 586
E +KKV+AKNSLENYAY R+ + K++E +E I+WLD NQLAEA+
Sbjct: 1715EHKKKVEAKNSLENYAYNMRNTIKDEKISSKLSGGDKKQIEDAIEGAIQWLDANQLAEAD 1894
Query: 587 EFEYKKQELE 596
EFE K +ELE
Sbjct: 1895EFEDKMKELE 1924
>TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chaperone
hsc70-3 - tomato, complete
Length = 2295
Score = 792 bits (2046), Expect = 0.0
Identities = 406/610 (66%), Positives = 488/610 (79%), Gaps = 14/610 (2%)
Frame = +1
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
MA + AIGIDLGTTYSCV VW H+RVEII N+ GNRTTPSYVAFTDTERL+GDAA N
Sbjct: 130 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 309
Query: 61 QLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAA 120
Q+A+NP NTVFDAKRLIGRR SD SVQ DMKLWPFKV+ G DKPMI V YK EE++ +A
Sbjct: 310 QVAMNPTNTVFDAKRLIGRRISDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQFSA 489
Query: 121 EEISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEP 180
EEISSMVL KMKE+AEAYLG +K AV+TVPAYFN++QRQ+TKDAG I+G +VMRIINEP
Sbjct: 490 EEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEP 669
Query: 181 TAASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFD 240
TAA+IAYGLDKK GE+NVL+FDLGGGTFDVSL+TI+EGIF+V AT GDT+LGG DFD
Sbjct: 670 TAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 849
Query: 241 NNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLH 300
N +VN V F+RK+ KDI+ N ++LRR+R ACE+AKR LSST+QTTIE+DSL GID +
Sbjct: 850 NRMVNHFVQEFKRKNKKDISGNPRALRRVRTACERAKRTLSSTAQTTIEIDSLFEGIDFY 1029
Query: 301 VTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMF 360
T+T+ FE+LN DLF KCME VEKCL +AK+ K+ V ++VLVGGSTRIPKVQQLL++ F
Sbjct: 1030TTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVDDVVLVGGSTRIPKVQQLLQDFF 1209
Query: 361 SVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVM 420
+ KELCKSINPDEAVAYGAAVQAAILS EG++KV++LLLLDV PLSLG+E GVM
Sbjct: 1210N----GKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVM 1377
Query: 421 SVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKG 480
+VLIP+NTTIPTKKE+ F T SDNQ V I+VYEGE +T N LGKFELSG PAP+G
Sbjct: 1378TVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRG 1557
Query: 481 DTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDE 540
+I VCFD+DA+GI+ VSAED + K ITITN GRLS+E++ +MV++A +YK+EDE
Sbjct: 1558VPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDE 1737
Query: 541 EVRKKVKAKNSLENYAYEARDRV--------------KKLEKMVEEVIEWLDRNQLAEAE 586
E ++KV+AKN+LENYAY R+ + KK+E +E I+WLD NQL EA+
Sbjct: 1738EHKRKVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEAD 1917
Query: 587 EFEYKKQELE 596
EFE K +ELE
Sbjct: 1918EFEDKMKELE 1947
>TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protein 4
precursor (BiP 4) (78 kDa glucose-regulated protein
homolog 4) (GRP 78-4)., partial (94%)
Length = 2265
Score = 632 bits (1629), Expect = 0.0
Identities = 330/610 (54%), Positives = 442/610 (72%), Gaps = 16/610 (2%)
Frame = +2
Query: 3 TSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAINQL 62
T+K IGIDLGTTYSCV V+ + VEII N+ GNR TPS+V+FTD ERL+G+AA N
Sbjct: 143 TTKLGTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLA 322
Query: 63 ALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYK-GEERKLAAE 121
A+NP T+FD KRLIGR+F+D+ VQ+DMKL P+K+V KP I V K GE + + E
Sbjct: 323 AVNPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIVNKD-GKPYIQVRVKDGETKVFSPE 499
Query: 122 EISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPT 181
E+S+M+L KMKE AEA+LG ++ AV+TVPAYFN+AQRQ+TKDAG IAG +V RIINEPT
Sbjct: 500 EVSAMILTKMKETAEAFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPT 679
Query: 182 AASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFDN 241
AA+IAYGLDKKG GE+N+LVFDLGGGTFDVS++TID G+F+V +T GDT+LGG DFD
Sbjct: 680 AAAIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLSTNGDTHLGGEDFDQ 850
Query: 242 NLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLHV 301
++ + + ++KH+KDI+++ ++L +LR E+AKR LSS Q +E++SL G+D
Sbjct: 851 RIMEYFIKLIKKKHSKDISKDNRALGKLRRESERAKRALSSQHQVRVEIESLFDGVDFSE 1030
Query: 302 TVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFS 361
+T+ FE+LN DLF K M V+K + +A + K+++ E+VLVGGSTRIPKVQQLLK+ F
Sbjct: 1031PLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFD 1210
Query: 362 VHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMS 421
KE K +NPDEAVA+GAAVQ +ILS EG ++ +++LLLDV PL+LGIE V GVM+
Sbjct: 1211G----KEPNKGVNPDEAVAFGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMT 1378
Query: 422 VLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGD 481
LIP+NT IPTKK + F T D QT+V I+V+EGE + T LG F+LSG PAP+G
Sbjct: 1379KLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGNFDLSGIPPAPRGT 1558
Query: 482 TEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEE 541
+I V F+VDA+GI+ V AED + ITITN GRLSQEE+ RMVR+A ++ ED++
Sbjct: 1559PQIEVTFEVDANGILNVRAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEEDKK 1738
Query: 542 VRKKVKAKNSLENYAYEARDRV---------------KKLEKMVEEVIEWLDRNQLAEAE 586
V++++ A+N+LE Y Y ++++ +K+E V+E +EWLD NQ E E
Sbjct: 1739VKERIDARNALETYVYNMKNQISDKDKLADKLESDEKEKIEAAVKEALEWLDDNQTVEKE 1918
Query: 587 EFEYKKQELE 596
EFE K +E+E
Sbjct: 1919EFEEKLKEVE 1948
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 486 bits (1252), Expect = e-138
Identities = 276/575 (48%), Positives = 378/575 (65%), Gaps = 6/575 (1%)
Frame = +2
Query: 10 IGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDT-ERLMGDAAINQLALNPHN 68
IGIDLGTT SCV+V +++ N G RTTPS VAFT E L+G A Q NP N
Sbjct: 263 IGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPEN 442
Query: 69 TVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAAEEISSMVL 128
T+ AKRLIGRRF D Q++MK+ P+K+V+ + KG++ + +I + VL
Sbjct: 443 TISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEA--KGQQ--YSPSQIGAFVL 610
Query: 129 FKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPTAASIAYG 188
KMKE AEAYLG V +AVITVPAYFN+AQRQ+TKDAG IAG +V+RIINEPTAA+++YG
Sbjct: 611 TKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYG 790
Query: 189 LDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFDNNLVNFLV 248
++K+G + VFDLGGGTFDVS++ I G+F+V AT GDT+LGG DFDN L++FLV
Sbjct: 791 MNKEGL------IAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 952
Query: 249 DFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLC----GGIDLHVTVT 304
+ F+R + D++++ +L+RLR A EKAK LSSTSQT I L + G L++T+T
Sbjct: 953 NEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLT 1132
Query: 305 QGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFSVHG 364
+ FE L +L E+ + CL +A I ++ E++LVGG TR+PKVQ+++ +F
Sbjct: 1133RSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFG--- 1303
Query: 365 RVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMSVLI 424
K CK +NPDEAVA GAA+Q IL + V+ELLLLDV PLSLGIE + G+ + LI
Sbjct: 1304--KSPCKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLI 1465
Query: 425 PKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDTEI 484
+NTTIPTKK + F T +DNQT V IKV +GE N LG+FEL G PAP+G +I
Sbjct: 1466NRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQI 1645
Query: 485 NVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEVRK 544
V FD+DA+GIV VSA+D S ++ ITI + G LS +E++ MV++A + +D+E +
Sbjct: 1646EVTFDIDANGIVTVSAKDKSTGKEQQITIKSS-GGLSDDEIQNMVKEAELHAQKDQERKS 1822
Query: 545 KVKAKNSLENYAYEARDRVKKL-EKMVEEVIEWLD 578
+ +NS + Y + + EK+ EV + ++
Sbjct: 1823LIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIE 1927
>TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chaperone PHSP1
precursor mitochondrial - garden pea, partial (98%)
Length = 2422
Score = 461 bits (1187), Expect = e-130
Identities = 268/579 (46%), Positives = 368/579 (63%), Gaps = 8/579 (1%)
Frame = +1
Query: 10 IGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDT-ERLMGDAAINQLALNPHN 68
IGIDLGTT SCV++ ++I N G RTTPS VAF E L+G A Q NP N
Sbjct: 247 IGIDLGTTNSCVSLMEGKNPKVIENSEGARTTPSVVAFNQKGELLVGTPAKRQAVTNPTN 426
Query: 69 TVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAAEEISSMVL 128
T+F KRLIGRRF D Q++MK+ P+K+V+ + + +++ + +I + VL
Sbjct: 427 TLFGTKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEIN----KQQYSPSQIGAFVL 594
Query: 129 FKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPTAASIAYG 188
KMKE AEAYLG + +AV+TVPAYFN+AQRQ+TKDAG IAG +V RIINEPTAA+++YG
Sbjct: 595 TKMKETAEAYLGKTISKAVVTVPAYFNDAQRQATKDAGRIAGLEVKRIINEPTAAALSYG 774
Query: 189 LDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFDNNLVNFLV 248
++ K E + VFDLGGGTFDVS++ I G+F+V AT GDT+LGG DFDN L++FLV
Sbjct: 775 MNNK-----EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 939
Query: 249 DFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSL----CGGIDLHVTVT 304
F+R + D+ ++ +L+RLR A EKAK LSSTSQT I L + G L++T+T
Sbjct: 940 SEFKRTDSIDLAKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLT 1119
Query: 305 QGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFSVHG 364
+ FE L +L E+ + CL +A I +V E++LVGG TR+PKVQ+++ E+F
Sbjct: 1120RSKFEALVNNLIERTKAPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQEVVSEIFG--- 1290
Query: 365 RVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMSVLI 424
K K +NPDEAVA GAA+Q IL + V+ELLLLDV PLSLGIE + G+ + LI
Sbjct: 1291--KSPSKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLI 1452
Query: 425 PKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAK--TDKNFFLGKFELSGFSPAPKGDT 482
+NTTIPTKK + F T +DNQT EG + N LG+F+L G PAP+G
Sbjct: 1453SRNTTIPTKKSQVFSTAADNQTQRGYXRCSQEGXREMAADNKSLGEFDLVGIPPAPRGLP 1632
Query: 483 EINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEV 542
+I V FD+DA+GIV VSA+D S ++ ITI + G LS +E+ MV++A + D+E
Sbjct: 1633QIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSS-GGLSDDEINNMVKEAELHAQRDQER 1809
Query: 543 RKKVKAKNSLENYAYEARDRVKKL-EKMVEEVIEWLDRN 580
+ + KNS + Y + + EK+ EV + ++ +
Sbjct: 1810KALIDIKNSADTSIYSIEKSLSEYREKIPSEVAKEIENS 1926
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 441 bits (1135), Expect = e-124
Identities = 220/287 (76%), Positives = 252/287 (87%)
Frame = +2
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
MAT K+ KAIGIDLGTTYSCV VW ++RVEIIPN+ GNRTTPSYVAFTDTERL+GDAA N
Sbjct: 98 MAT-KEGKAIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKN 274
Query: 61 QLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAA 120
Q+A+NP NTVFDAKRLIGRRFSD SVQ DMKLWPFKVV G +KPMI V YKGEE+K AA
Sbjct: 275 QVAMNPQNTVFDAKRLIGRRFSDESVQNDMKLWPFKVVPGPAEKPMIVVNYKGEEKKFAA 454
Query: 121 EEISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEP 180
EEISSMVL KM+EVAEA+LGH VK AV+TVPAYFN++QRQ+TKDAG I+G +V+RIINEP
Sbjct: 455 EEISSMVLIKMREVAEAFLGHPVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEP 634
Query: 181 TAASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFD 240
TAA+IAYGLDKK RKGEQNVL+FDLGGGTFDVSL+TI+EGIF+V AT GDT+LGG DFD
Sbjct: 635 TAAAIAYGLDKKASRKGEQNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 814
Query: 241 NNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTT 287
N +VN V F+RK+ KDI+ N ++LRRLR ACE+AKR LSST+QTT
Sbjct: 815 NRMVNHFVSEFRRKNKKDISGNARALRRLRTACERAKRTLSSTAQTT 955
>TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa protein
(HSP70). {Neurospora crassa}, partial (54%)
Length = 1258
Score = 433 bits (1113), Expect(2) = e-124
Identities = 222/341 (65%), Positives = 277/341 (81%)
Frame = +2
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
MATS AIGIDLGTTYSCV + ++++EII N+ GNRTTPSYVAF DTERL+GDAA N
Sbjct: 173 MATSF---AIGIDLGTTYSCVGRYANDKIEIIANDQGNRTTPSYVAFNDTERLIGDAAKN 343
Query: 61 QLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAA 120
Q+A+NPHNTVFDAKRLIGR+FSD VQ DMK +PFKV+ KP I V +KGE +
Sbjct: 344 QVAMNPHNTVFDAKRLIGRKFSDSEVQADMKHFPFKVID-KGGKPNIEVEFKGENKTFTP 520
Query: 121 EEISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEP 180
EEIS+MVL KM+E AEAYLG +V AVITVPAYFN++QRQ+TKDAG IAG +V+RIINEP
Sbjct: 521 EEISAMVLVKMRETAEAYLGGQVTNAVITVPAYFNDSQRQATKDAGLIAGLNVLRIINEP 700
Query: 181 TAASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFD 240
TAA+IAYGLDKK +GE+NVL+FDLGGGTFDVSL+TI+EGIF+V +T GDT+LGG DFD
Sbjct: 701 TAAAIAYGLDKKA--EGERNVLIFDLGGGTFDVSLLTIEEGIFEVKSTAGDTHLGGEDFD 874
Query: 241 NNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLH 300
N LVN V+ F+RKH KD++ N ++LRRLR ACE+AKR LSS++QT+IE+DSL GID +
Sbjct: 875 NRLVNHFVNEFKRKHKKDLSSNARALRRLRTACERAKRTLSSSAQTSIEIDSLFEGIDFY 1054
Query: 301 VTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELV 341
++T+ FE+L +DLF ++ V++ L +AKI KS+VHE+V
Sbjct: 1055TSITRARFEELCQDLFRSTIQPVDRVLSDAKIDKSQVHEIV 1177
Score = 30.0 bits (66), Expect(2) = e-124
Identities = 11/19 (57%), Positives = 17/19 (88%)
Frame = +3
Query: 343 VGGSTRIPKVQQLLKEMFS 361
VGGSTRIP++Q+L+ + F+
Sbjct: 1182 VGGSTRIPRIQKLISDYFN 1238
>TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chaperone
HSC71.0 - garden pea, partial (59%)
Length = 1336
Score = 412 bits (1060), Expect = e-115
Identities = 216/347 (62%), Positives = 267/347 (76%), Gaps = 14/347 (4%)
Frame = +2
Query: 264 KSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLHVTVTQGLFEKLNKDLFEKCMEIV 323
++LRRLR ACE+AKR LSST+QTTIE+DSL GID + +T+ FE+LN DLF KCME V
Sbjct: 2 RALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFYSPITRARFEELNMDLFRKCMEPV 181
Query: 324 EKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFSVHGRVKELCKSINPDEAVAYGA 383
EKCL +AK+ K VH++VLVGGSTRIPKVQQLL++ F+ KELCKSINPDEAVAYGA
Sbjct: 182 EKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQLLQDFFNG----KELCKSINPDEAVAYGA 349
Query: 384 AVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMSVLIPKNTTIPTKKERTFRTTSD 443
AVQAAILS EG++KV++LLLLDV PLSLG+E GVM+VLIP+NTTIPTKKE+ F T SD
Sbjct: 350 AVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSD 529
Query: 444 NQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAEDM 503
NQ V I+V+EGE +T N LGKFELSG PAP+G +I VCFD+DA+GI+ VSAED
Sbjct: 530 NQPGVLIQVFEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDK 709
Query: 504 SLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDRV 563
+ K ITITN GRLS+E++ +MV++A +YK+EDEE +KKV+AKN+LENYAY R+ +
Sbjct: 710 TTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTI 889
Query: 564 --------------KKLEKMVEEVIEWLDRNQLAEAEEFEYKKQELE 596
KK+E +E I+WLD NQLAEA+EFE K +ELE
Sbjct: 890 KDEKIAGKLDSDDKKKIEDTIEAAIQWLDANQLAEADEFEDKMKELE 1030
>TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (53%)
Length = 1189
Score = 379 bits (974), Expect = e-105
Identities = 197/308 (63%), Positives = 241/308 (77%), Gaps = 14/308 (4%)
Frame = +3
Query: 303 VTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFSV 362
+T+ FE+LN DLF KCME VEKCL +AKI KS VHE+VLVGGSTRIPKVQQLL++ F+
Sbjct: 3 ITRARFEELNMDLFRKCMEPVEKCLRDAKIDKSHVHEVVLVGGSTRIPKVQQLLQDFFNG 182
Query: 363 HGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMSV 422
KELCKSINPDEAVAYGAAVQAAILS EGD+KV++LLLLDV PLSLG+E GVM+
Sbjct: 183 ----KELCKSINPDEAVAYGAAVQAAILSGEGDEKVQDLLLLDVTPLSLGLETAGGVMTT 350
Query: 423 LIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDT 482
LIP+NTTIPTKKE+ F T SDNQ V I+V+EGE A+T N LGKFEL+G PAP+G
Sbjct: 351 LIPRNTTIPTKKEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPRGVP 530
Query: 483 EINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEV 542
+INVCFD+DA+GI+ VSAED + +K ITITN GRLS+EE+ +MV+DA +YKAEDEEV
Sbjct: 531 QINVCFDIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAEKYKAEDEEV 710
Query: 543 RKKVKAKNSLENYAYEARDRVK--------------KLEKMVEEVIEWLDRNQLAEAEEF 588
+KKV+AKNS+ENYAY R+ +K K+EK VE+ I+WL+ NQ+AE +EF
Sbjct: 711 KKKVEAKNSIENYAYNMRNTIKDEKIGGKLSHEDKEKIEKAVEDAIQWLEGNQMAEVDEF 890
Query: 589 EYKKQELE 596
E K++ELE
Sbjct: 891 EDKQKELE 914
>TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chaperone BiP-A
- soybean, partial (52%)
Length = 1231
Score = 295 bits (755), Expect = 3e-80
Identities = 158/316 (50%), Positives = 215/316 (68%), Gaps = 15/316 (4%)
Frame = +1
Query: 296 GIDLHVTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQL 355
G+ +T+ FE+LN DLF K M V+K + +A + K+++ E+VLVGGSTRIPKVQQL
Sbjct: 4 GVAFSEPLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQL 183
Query: 356 LKEMFSVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEV 415
LK+ F KE K +NPDEAVA+GAAVQ +ILS EG + +++LLLDV PL+LGIE
Sbjct: 184 LKDYFDG----KEPNKGVNPDEAVAFGAAVQGSILSGEGGAETKDILLLDVAPLTLGIET 351
Query: 416 VEGVMSVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFS 475
V GVM+ LIP+NT IPTKK + F T D QT+V I+V+EGE + T LGKF+LSG
Sbjct: 352 VGGVMTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIP 531
Query: 476 PAPKGDTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQY 535
PAP+G +I V F+VDA+GI+ V AED + ITITN GRLSQEE+ RMVR+A ++
Sbjct: 532 PAPRGTPQIEVTFEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEF 711
Query: 536 KAEDEEVRKKVKAKNSLENYAYEARDRV---------------KKLEKMVEEVIEWLDRN 580
ED++V++++ A+N+LE Y Y +++V +K+E V+E +EWLD N
Sbjct: 712 AEEDKKVKERIDARNALETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDDN 891
Query: 581 QLAEAEEFEYKKQELE 596
Q E EE+E K +E+E
Sbjct: 892 QSVEKEEYEEKLKEVE 939
>BQ139103 homologue to GP|2655420|gb| heat shock cognate protein HSC70
{Brassica napus}, partial (29%)
Length = 628
Score = 295 bits (754), Expect = 4e-80
Identities = 143/191 (74%), Positives = 166/191 (86%)
Frame = +2
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
MA + AIGIDLGTTYSCV VW H+RVEII N+ GNRTTPSYVAFTD+ERL+GDAA N
Sbjct: 56 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 235
Query: 61 QLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAA 120
Q+A+NP NTVFDAKRLIGRRFSD SVQ DMKLWPFK++ G +KP+I V YKGE+++ AA
Sbjct: 236 QVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKIISGPAEKPLIGVNYKGEDKEFAA 415
Query: 121 EEISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEP 180
EEISSMVL KM+E+AEAYLG +K AV+TVPAYFN++QRQ+TKDAG IAG +VMRIINEP
Sbjct: 416 EEISSMVLMKMREIAEAYLGSAIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEP 595
Query: 181 TAASIAYGLDK 191
TAA+IAYGLDK
Sbjct: 596 TAAAIAYGLDK 628
>TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic reticulum
HSC70-cognate binding protein precursor {Glycine max},
partial (38%)
Length = 962
Score = 285 bits (729), Expect = 4e-77
Identities = 149/232 (64%), Positives = 183/232 (78%), Gaps = 2/232 (0%)
Frame = +1
Query: 4 SKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAINQLA 63
+K IGIDLGTTYSCV V+ + VEII N+ GNR TPS+V+FTD ERL+G+AA N A
Sbjct: 280 TKLGTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAA 459
Query: 64 LNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRD-KPMISVTYK-GEERKLAAE 121
+NP T+FD KRLIGR+F D+ VQ+DMKL P+K+V RD KP I V K GE + + E
Sbjct: 460 VNPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIV--NRDGKPYIQVRVKDGETKVFSPE 633
Query: 122 EISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPT 181
EIS+M+L KMKE AE +LG ++ AV+TVPAYFN+AQRQ+TKDAG IAG +V RIINEPT
Sbjct: 634 EISAMILGKMKETAEGFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPT 813
Query: 182 AASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTY 233
AA+IAYGLDKKG GE+N+LVFDLGGGTFDVS++TID G+F+V AT GDT+
Sbjct: 814 AAAIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTH 960
>TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chaperone hsc70-3
- tomato, partial (27%)
Length = 667
Score = 254 bits (648), Expect = 9e-68
Identities = 128/192 (66%), Positives = 152/192 (78%)
Frame = +3
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
M+ K AIGIDLGTTYSCVAV + ++II N+LG RTTPS+VAF D+ER++GDAA N
Sbjct: 81 MSGRGKKVAIGIDLGTTYSCVAVCKNGEIDIIVNDLGKRTTPSFVAFKDSERMIGDAAFN 260
Query: 61 QLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAA 120
A NP NT+FDAKRLIGR+FSD VQ D+KLWPFKV+ DKPMI V Y EE+ AA
Sbjct: 261 IAASNPTNTIFDAKRLIGRKFSDPIVQSDVKLWPFKVIGDLNDKPMIVVNYNDEEKHFAA 440
Query: 121 EEISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEP 180
EEISSMVL KM+E+AE +LG V+ VITVPAYFN++QRQST+DAG IAG +VMRIINEP
Sbjct: 441 EEISSMVLVKMREIAETFLGSIVEDVVITVPAYFNDSQRQSTRDAGAIAGLNVMRIINEP 620
Query: 181 TAASIAYGLDKK 192
TAA+IAYG + K
Sbjct: 621 TAAAIAYGFNTK 656
>TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [imported] -
Arabidopsis thaliana, partial (91%)
Length = 1717
Score = 164 bits (414), Expect(2) = 4e-65
Identities = 88/239 (36%), Positives = 142/239 (58%), Gaps = 1/239 (0%)
Frame = +1
Query: 261 ENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLHVTVTQGLFEKLNKDLFEKCM 320
E +KS+ LR+A + A LS+ S +++D L + + V + FE++NK++FEKC
Sbjct: 763 EEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCE 939
Query: 321 EIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFSVHGRVKELCKSINPDEAVA 380
++ +CL +AK+ ++++++LVGG + IP+V+ L+ + ++ E+ K INP E
Sbjct: 940 SLIIQCLHDAKVEVGDINDVILVGGCSYIPRVENLVTNLC----KITEVYKGINPLEGAV 1107
Query: 381 YGAAVQAAILSSEGDKKVE-ELLLLDVMPLSLGIEVVEGVMSVLIPKNTTIPTKKERTFR 439
GA ++ A+ S D +LL + PL++GI +IP+NTT+P +K+ F
Sbjct: 1108CGATMEGAVASGISDPFGNLDLLTIQATPLAIGIRADGNKFVPVIPRNTTMPARKDLLFT 1287
Query: 440 TTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEV 498
T DNQT I VYEGEG K ++N LG F++ G APKG EI+VC D+DA ++ V
Sbjct: 1288TIHDNQTEALILVYEGEGKKAEENHLLGYFKIMGIPAAPKGVPEISVCMDIDAANVLRV 1464
Score = 103 bits (256), Expect(2) = 4e-65
Identities = 73/245 (29%), Positives = 124/245 (49%), Gaps = 11/245 (4%)
Frame = +3
Query: 15 GTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAA--INQLALNPHNTVFD 72
GT+ VAVWN + VE++ N+ + S+V F D G + ++ + +T+F+
Sbjct: 3 GTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFGDTIFN 182
Query: 73 AKRLIGRRFSDRSVQQDMKLWPFKVVQGTRD---KPMISVTYKGEERKLAAEEISSMVLF 129
KRLIGR +D V L PF V T D +P I+ R EE+ +M L
Sbjct: 183 MKRLIGRVDTDPVVHASKNL-PFLVQ--TLDIGVRPFIAALVNNVWRSTTPEEVLAMFLV 353
Query: 130 KMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPTAASIAYGL 189
+++ + E +L ++ V+TVP F+ Q + A +AG V+R++ EPTA ++ YG
Sbjct: 354 ELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALLYGQ 533
Query: 190 DKKGWRK------GEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFDNNL 243
++ + E+ L+F++G G DV++ G+ ++ A G T +GG D N+
Sbjct: 534 QQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLLQNM 710
Query: 244 VNFLV 248
+ L+
Sbjct: 711 MRHLL 725
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 239 bits (611), Expect = 2e-63
Identities = 162/542 (29%), Positives = 264/542 (47%), Gaps = 14/542 (2%)
Frame = +1
Query: 6 KDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAINQLALN 65
K +G D G VAV ++++ N+ R TP+ V F D +R +G A +N
Sbjct: 235 KMSVVGFDFGNESCIVAVARQRGIDVVLNDESKRETPAIVCFGDKQRFIGTAGAASTMMN 414
Query: 66 PHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAAEEISS 125
P N++ KRLIG++F+D +Q+D+K PF V +G P+I Y GE R+ A ++
Sbjct: 415 PKNSISQIKRLIGKKFADPELQRDLKSLPFNVTEGPDGYPLIHARYLGESREFTATQVFG 594
Query: 126 MVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPTAASI 185
M+L +KE+A+ L V I +P YF + QR+S DA IAG + +I+E TA ++
Sbjct: 595 MMLSNLKEIAQKNLNAAVVDCCIGIPVYFTDLQRRSVLDAATIAGLHPLHLIHETTATAL 774
Query: 186 AYGLDKKGWRKGE-QNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFDNNLV 244
AYG+ K + E NV D+G + V + +G V + D LGG DFD L
Sbjct: 775 AYGIYKTDLPENEWLNVAFVDVGHASMQVCIAGFKKGQLHVLSHSYDRSLGGRDFDEALF 954
Query: 245 NFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLHVTVT 304
+ F+ ++ D+ +N ++ RLR ACEK K++LS+ + + ++ L D+ +
Sbjct: 955 HHFAAKFKEEYKIDVYQNARACLRLRAACEKLKKVLSANPEAPLNIECLMDEKDVRGFIK 1134
Query: 305 QGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFSVHG 364
+ FE+L+ + E+ +EK L EA + +H + +VG +R+P + ++L E F
Sbjct: 1135RDDFEQLSLPILERVKGPLEKALAEAGLTVENIHMVEVVGSGSRVPAINKILTEFFK--- 1305
Query: 365 RVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGI----------- 413
KE +++N E VA GAA+Q AILS KV E + + P S+ +
Sbjct: 1306--KEPRRTMNASECVARGAALQCAILSP--TFKVREFQVNESFPFSVSLSWKYSGSDAPD 1473
Query: 414 -EVVEGVMSVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYE-GEGAKTDKNFFLGKFEL 471
E +++ PK IP+ K TF T SV ++ ++ E + +G F+
Sbjct: 1474SESDNKQSTIVFPKGNPIPSSKVLTFFRT--GTFSVDVQCHDLSETPTKISTYTIGPFQT 1647
Query: 472 SGFSPAPKGDTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRD 531
KG + V ++ GIV V + + + + +T EE +M D
Sbjct: 1648KN---GDKGKVKAKV--RLNLHGIVSVESATLFEEEEIEVPVTKEFA----EENAKMETD 1800
Query: 532 AA 533
A
Sbjct: 1801EA 1806
>TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protein 70
{Cucumis sativus}, partial (35%)
Length = 723
Score = 236 bits (603), Expect = 1e-62
Identities = 123/212 (58%), Positives = 154/212 (72%), Gaps = 14/212 (6%)
Frame = +3
Query: 399 EELLLLDVMPLSLGIEVVEGVMSVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGA 458
++LLLLDV PLSLG+E GVM+VLIP+NTTIPTKKE+ F T SDNQ V I+VYEGE A
Sbjct: 3 QDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERA 182
Query: 459 KTDKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLG 518
+T N LGKFELSG PAP+G +I VCFD+DA+GI+ VSAED + K ITITN G
Sbjct: 183 RTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKG 362
Query: 519 RLSQEEMRRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDRV--------------K 564
RLS+EE+ +MV +A +YKAEDEE +KKV AKN+LENYAY R+ + K
Sbjct: 363 RLSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKK 542
Query: 565 KLEKMVEEVIEWLDRNQLAEAEEFEYKKQELE 596
K++ ++ I+WLD NQLAEA+EF+ K +ELE
Sbjct: 543 KIDDAIDAAIQWLDSNQLAEADEFQDKMKELE 638
>BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated protein
homolog precursor (GRP 78), partial (47%)
Length = 1032
Score = 219 bits (557), Expect(2) = 2e-61
Identities = 122/263 (46%), Positives = 175/263 (66%), Gaps = 16/263 (6%)
Frame = -1
Query: 349 IPKVQQLLKEMFSVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMP 408
IPKVQ L++E F K+ K INPDEAVA+GAAVQA +LS G++ EE++L+DV P
Sbjct: 900 IPKVQSLIEEYFGG----KKASKGINPDEAVAFGAAVQAGVLS--GEEGTEEIVLMDVNP 739
Query: 409 LSLGIEVVEGVMSVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGK 468
L+LGIE GVM+ LI +NT IPT+K + F T +DNQ V I+V+EGE + T N LGK
Sbjct: 738 LTLGIETTGGVMTKLITRNTPIPTRKSQIFSTAADNQPVVLIQVFEGERSLTKDNNQLGK 559
Query: 469 FELSGFSPAPKGDTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRM 528
FEL+G PAP+G +I V F++DA+GI++VSA D +++ITITN GRL++EE+ RM
Sbjct: 558 FELTGIPPAPRGVPQIEVSFELDANGILKVSAHDKGTGKQESITITNDKGRLTKEEIDRM 379
Query: 529 VRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDRVKKLE---------------KMVEEV 573
V +A +Y ED+ R++++A+N LENYA+ +++V E + V+E
Sbjct: 378 VEEAEKYAEEDKATRERIEARNGLENYAFSLKNQVNDEEGLGGKIDDEDKETILEAVKET 199
Query: 574 IEWLDRN-QLAEAEEFEYKKQEL 595
+WL+ N A E+FE +K++L
Sbjct: 198 NDWLEENGATANTEDFEEQKEKL 130
Score = 36.2 bits (82), Expect(2) = 2e-61
Identities = 16/29 (55%), Positives = 23/29 (79%)
Frame = -3
Query: 323 VEKCLVEAKIGKSEVHELVLVGGSTRIPK 351
VE+ L +AK+ K +V ++VLVGGSTR P+
Sbjct: 979 VEQVLKDAKVKKEDVDDIVLVGGSTRYPQ 893
>AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 kDa protein.
[Petunia] {Petunia hybrida}, partial (24%)
Length = 532
Score = 211 bits (538), Expect = 5e-55
Identities = 109/167 (65%), Positives = 133/167 (79%), Gaps = 2/167 (1%)
Frame = +1
Query: 1 MATSKKDKAIGIDLGTTYSCVAVW--NHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAA 58
MA + + A+GIDLGTTYSCVAVW NRVEII N+ G++ TPS+VAFTD +RL+G AA
Sbjct: 31 MANNCECYAVGIDLGTTYSCVAVWLEEKNRVEIIHNDEGSKITPSFVAFTDGQRLVGAAA 210
Query: 59 INQLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKL 118
+Q A+NP NTVFDAKRLIGR+FSD VQ+D+ LWPFKVV G DKPMIS+ +KG+E+ L
Sbjct: 211 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDILLWPFKVVSGVNDKPMISLKFKGQEKLL 390
Query: 119 AAEEISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDA 165
AEEISS+VL M+E+AE YL VK A ITVPAYFN+AQR++T DA
Sbjct: 391 CAEEISSIVLTNMREIAEMYLESPVKNAGITVPAYFNDAQRKATIDA 531
>CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus stolonifer},
partial (19%)
Length = 527
Score = 181 bits (459), Expect = 7e-46
Identities = 89/126 (70%), Positives = 104/126 (81%)
Frame = +3
Query: 5 KKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAINQLAL 64
++ KA+GIDLGTTYSCV VW ++RVEII N+ GNRTTPSYVAFTDTERL+GDAA NQ+A+
Sbjct: 153 QEGKAVGIDLGTTYSCVGVWQNDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAM 332
Query: 65 NPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAAEEIS 124
NP+NTVFDAKRLIGRRF+D+ VQ DMK WPFKV+ KP I V YKGE ++ EEIS
Sbjct: 333 NPYNTVFDAKRLIGRRFADQEVQSDMKHWPFKVIDKAA-KPYIQVEYKGETKQFTPEEIS 509
Query: 125 SMVLFK 130
SMVL K
Sbjct: 510 SMVLTK 527
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,845,676
Number of Sequences: 36976
Number of extensions: 175083
Number of successful extensions: 917
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 855
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 862
length of query: 602
length of database: 9,014,727
effective HSP length: 102
effective length of query: 500
effective length of database: 5,243,175
effective search space: 2621587500
effective search space used: 2621587500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0118c.15


