

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0118a.3
(328 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
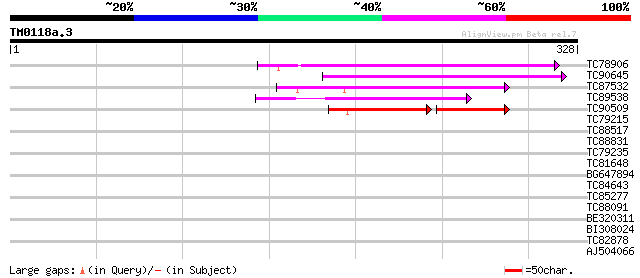
Score E
Sequences producing significant alignments: (bits) Value
TC78906 similar to GP|18377616|gb|AAL66958.1 unknown protein {Ar... 118 4e-27
TC90645 weakly similar to GP|15289850|dbj|BAB63547. contains EST... 103 9e-23
TC87532 similar to GP|21537285|gb|AAM61626.1 unknown {Arabidopsi... 80 1e-15
TC89538 weakly similar to GP|21689895|gb|AAM67508.1 unknown prot... 67 9e-12
TC90509 similar to GP|21537285|gb|AAM61626.1 unknown {Arabidopsi... 49 3e-06
TC79215 similar to GP|20259431|gb|AAM14036.1 unknown protein {Ar... 32 0.26
TC88517 similar to GP|19386853|dbj|BAB86231. hypothetical protei... 32 0.34
TC88831 similar to GP|9758871|dbj|BAB09425.1 gb|AAD20092.1~gene_... 31 0.75
TC79235 weakly similar to GP|7297741|gb|AAF52992.1| CG17108 gene... 29 2.9
TC81648 similar to PIR|T00840|T00840 probable senescence-related... 29 2.9
BG647894 similar to GP|17381274|gb AT5g11700/T22P22_90 {Arabidop... 28 3.7
TC84643 similar to GP|21689887|gb|AAM67504.1 unknown protein {Ar... 28 3.7
TC85277 similar to GP|15293299|gb|AAK93760.1 unknown protein {Ar... 28 6.4
TC88091 homologue to SP|P51819|HS83_PHANI Heat shock protein 83.... 28 6.4
BE320311 similar to PIR|G84781|G847 hypothetical protein At2g365... 28 6.4
BI308024 28 6.4
TC82878 similar to PIR|T45677|T45677 ATP-dependent RNA helicase-... 27 8.3
AJ504066 weakly similar to PIR|T37811|T378 very hypothetical pro... 27 8.3
>TC78906 similar to GP|18377616|gb|AAL66958.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1746
Score = 118 bits (295), Expect = 4e-27
Identities = 74/179 (41%), Positives = 103/179 (57%), Gaps = 4/179 (2%)
Frame = +2
Query: 144 PLLPPSISHRR---TVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVD 200
P L P S RR T+ LDLDETLVHS T + DF + YVK+RP +
Sbjct: 1013 PTLAPKQSPRRKSVTLVLDLDETLVHS-TLEHCDDADFTFNIFFNMKDYIVYVKQRPFLH 1189
Query: 201 EFLEALAAKFEVVVFTAALREYASLVLDRVDRN-RLVSHRLYRDSCKQEDGKLVKDLSLA 259
+FLE ++ FEVV+FTA+ YA+ +LD +D + + +S RLYR+SC DG KDL++
Sbjct: 1190 KFLERVSDMFEVVIFTASQSIYANQLLDILDPDEKFISRRLYRESCMFSDGNYTKDLTIL 1369
Query: 260 GRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRDAV 318
G DL +VVI+D++P F Q N I I+ + DD D L L F + + DD+R +
Sbjct: 1370 GIDLAKVVIIDNSPQVFRLQVNNGIPIKSWFDDPSDCALMSLLPFLETLADADDVRPII 1546
>TC90645 weakly similar to GP|15289850|dbj|BAB63547. contains ESTs
AU092190(C11346) AU062476(C11346)
AU063421(C61571)~unknown protein, partial (25%)
Length = 683
Score = 103 bits (257), Expect = 9e-23
Identities = 59/142 (41%), Positives = 84/142 (58%), Gaps = 1/142 (0%)
Frame = +3
Query: 182 PVIDGVPMEFYVKKRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNR-LVSHRL 240
PV+ YV+KRP + EFLE ++ FE+V+FTA+ + YA +LD +D ++ L S RL
Sbjct: 6 PVLLDKEYTVYVRKRPFLHEFLERVSKMFEIVIFTASKKIYAEKLLDVLDPDKKLFSRRL 185
Query: 241 YRDSCKQEDGKLVKDLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWK 300
YRDSC +DG KDL++ G DL +V IVD++P F Q N I I + DD D L
Sbjct: 186 YRDSCIYQDGTFTKDLTVLGIDLAKVAIVDNSPQVFRLQVNNGIPIESWFDDPSDSALIS 365
Query: 301 LRMFFDGVDCCDDMRDAVKQFF 322
L F + + DD+R + + F
Sbjct: 366 LLPFLEKLVDVDDVRPIIAEKF 431
>TC87532 similar to GP|21537285|gb|AAM61626.1 unknown {Arabidopsis
thaliana}, partial (84%)
Length = 1411
Score = 80.1 bits (196), Expect = 1e-15
Identities = 58/154 (37%), Positives = 79/154 (50%), Gaps = 19/154 (12%)
Frame = +3
Query: 155 TVFLDLDETLV--HSNTAPPPESFDFVVRPVIDGVPMEFY----------------VKKR 196
TV LDLDETLV + ++ P + ++ ME V +R
Sbjct: 528 TVVLDLDETLVCAYETSSLPAALRSQAIEAGLNWFDMECVSSDKEGEGRPKINYVTVFER 707
Query: 197 PGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDS-CKQEDGKLVKD 255
PG+ EFL L+ ++V+FTA L YA ++D +D+ S RLYR S E + VKD
Sbjct: 708 PGLKEFLTKLSEFADLVLFTAGLEGYARPLVDIIDKENRFSLRLYRPSTISTEHREHVKD 887
Query: 256 LSLAGRDLKRVVIVDDNPNSFANQPENAILIRPF 289
L+ +DL R+VIVD+NP SF QP N I PF
Sbjct: 888 LTCISKDLDRIVIVDNNPFSFVLQPVNGIPCIPF 989
>TC89538 weakly similar to GP|21689895|gb|AAM67508.1 unknown protein
{Arabidopsis thaliana}, partial (37%)
Length = 990
Score = 67.0 bits (162), Expect = 9e-12
Identities = 44/125 (35%), Positives = 62/125 (49%)
Frame = +3
Query: 143 LPLLPPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEF 202
LP L P H T+ LDL+ETL+H I + KRPGVD F
Sbjct: 630 LPDLLPQEQHVFTLVLDLNETLIH----------------YIWTRDTSWQTFKRPGVDAF 761
Query: 203 LEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRD 262
LE LA FE+ V+T + V++R+D + +RL R + K +DGK +DLS R+
Sbjct: 762 LEHLAQFFEIXVYTDEQNMFVDPVIERLDPKHCIXYRLSRPATKYQDGKHYRDLSKLNRN 941
Query: 263 LKRVV 267
+V+
Sbjct: 942 PAKVM 956
>TC90509 similar to GP|21537285|gb|AAM61626.1 unknown {Arabidopsis
thaliana}, partial (62%)
Length = 1317
Score = 48.5 bits (114), Expect = 3e-06
Identities = 27/62 (43%), Positives = 39/62 (62%), Gaps = 2/62 (3%)
Frame = +3
Query: 185 DGVPMEFYVK--KRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYR 242
DG P+ YV +RPG+ EFL+ L+ ++V+FTA + YA ++DR+D S RLYR
Sbjct: 303 DGEPIINYVTVFERPGLKEFLKQLSEFADLVLFTAGVEGYARPLVDRIDTENWFSLRLYR 482
Query: 243 DS 244
S
Sbjct: 483 PS 488
Score = 42.4 bits (98), Expect = 2e-04
Identities = 21/42 (50%), Positives = 27/42 (64%)
Frame = +2
Query: 248 EDGKLVKDLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPF 289
E + VKDL+ +DL +VIVD+NP SF QP+N I PF
Sbjct: 674 EHREYVKDLTCISKDLCNIVIVDNNPYSFLLQPDNGIPCVPF 799
>TC79215 similar to GP|20259431|gb|AAM14036.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1240
Score = 32.3 bits (72), Expect = 0.26
Identities = 29/118 (24%), Positives = 47/118 (39%), Gaps = 2/118 (1%)
Frame = +1
Query: 162 ETLVHSNTAPPPESFDFVVRPVIDGVPMEF--YVKKRPGVDEFLEALAAKFEVVVFTAAL 219
+ L ++ P P+ F P + G P Y++ R +D L LA + ++
Sbjct: 304 QALGNNTNVPVPKVFCLCNDPAVIGTPFYIMEYLEGRIFIDPKLPGLAPQSRSTIYRETA 483
Query: 220 REYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRDLKRVVIVDDNPNSFA 277
+ ASL VD L ++ D CK++ + K + D K NP FA
Sbjct: 484 KTLASLHSANVDSIGLGNYGRRNDYCKRQIERWAKQYVSSTSDGKPA----SNPKMFA 645
>TC88517 similar to GP|19386853|dbj|BAB86231. hypothetical protein~similar
to Homo sapiens chromosome 5 LOC134510, partial (70%)
Length = 1396
Score = 32.0 bits (71), Expect = 0.34
Identities = 38/190 (20%), Positives = 77/190 (40%), Gaps = 23/190 (12%)
Frame = +3
Query: 148 PSISHRRTVFLDLDETLV-HSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEFLEAL 206
P ++ + LD+D TL H +TA P RP + EFL A
Sbjct: 423 PCRQGKKLLVLDIDYTLFDHRSTAENPLQL------------------MRPYLHEFLTAA 548
Query: 207 AAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLV------------K 254
A++++++++A ++ +L + ++ ++ + Y+ + + ++ K
Sbjct: 549 YAEYDIMIWSATSMKWITLKMSQLG---VLDNPNYKITALLDHMGMITVQTPSRGVFDCK 719
Query: 255 DLSLAGRDL------KRVVIVDDNPNSFANQPENAILIRPF----VDDVFDRELWKLRMF 304
L L ++ DD +F P+N + I+PF + D+EL KL +
Sbjct: 720 PLGLIWAQFPEFYSASNTIMFDDLRRNFVMNPQNGLTIKPFKKAHANRDTDQELVKLTQY 899
Query: 305 FDGVDCCDDM 314
+ DD+
Sbjct: 900 LLAIAELDDL 929
>TC88831 similar to GP|9758871|dbj|BAB09425.1
gb|AAD20092.1~gene_id:K24G6.5~similar to unknown protein
{Arabidopsis thaliana}, partial (7%)
Length = 880
Score = 30.8 bits (68), Expect = 0.75
Identities = 19/52 (36%), Positives = 29/52 (55%), Gaps = 5/52 (9%)
Frame = -2
Query: 30 IFSSLRFLPYPNTKNQAMVSRVKRVP----TKSIKD-IRRNNNRRWRRKTPI 76
+ ++ FLP P ++ V V + T S K+ + RNNN R+RR+TPI
Sbjct: 195 LLETITFLPMPIPRSLVFVIVVVVIHLCGNTASTKEALMRNNNNRYRRETPI 40
>TC79235 weakly similar to GP|7297741|gb|AAF52992.1| CG17108 gene product
{Drosophila melanogaster}, partial (21%)
Length = 648
Score = 28.9 bits (63), Expect = 2.9
Identities = 13/35 (37%), Positives = 17/35 (48%)
Frame = -2
Query: 139 PSPPLPLLPPSISHRRTVFLDLDETLVHSNTAPPP 173
P PP P PP + H T +H++ APPP
Sbjct: 368 PPPPNPYAPPPVVHHH-------HTPLHAHPAPPP 285
>TC81648 similar to PIR|T00840|T00840 probable senescence-related protein
[imported] - Arabidopsis thaliana, partial (36%)
Length = 854
Score = 28.9 bits (63), Expect = 2.9
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = +2
Query: 139 PSPPLPLLPPSISHRRTVFLDLDETLVHSNTAPPP 173
PS PL LLP IS +++ + + S T PPP
Sbjct: 62 PSQPLLLLPNQISTLQSITISTTSLKISSLTPPPP 166
>BG647894 similar to GP|17381274|gb AT5g11700/T22P22_90 {Arabidopsis
thaliana}, partial (43%)
Length = 604
Score = 28.5 bits (62), Expect = 3.7
Identities = 19/50 (38%), Positives = 25/50 (50%), Gaps = 4/50 (8%)
Frame = -3
Query: 139 PSPPLPLLPPS--ISHRRTVFLDLDETLVHSNTAPPPE--SFDFVVRPVI 184
P PP PL+PPS ++ R + T H PPP SF VV P++
Sbjct: 425 PEPPPPLIPPSPPLAKRSPETSTVSTTSNHILPPPPPP*LSFLTVVPPLL 276
>TC84643 similar to GP|21689887|gb|AAM67504.1 unknown protein {Arabidopsis
thaliana}, partial (4%)
Length = 491
Score = 28.5 bits (62), Expect = 3.7
Identities = 15/35 (42%), Positives = 21/35 (59%)
Frame = -3
Query: 33 SLRFLPYPNTKNQAMVSRVKRVPTKSIKDIRRNNN 67
S++FL PN K ++ KRVP I+D+ NNN
Sbjct: 291 SIQFLNTPNLKTESF--HQKRVPI*RIRDLNCNNN 193
>TC85277 similar to GP|15293299|gb|AAK93760.1 unknown protein {Arabidopsis
thaliana}, partial (40%)
Length = 1976
Score = 27.7 bits (60), Expect = 6.4
Identities = 17/55 (30%), Positives = 25/55 (44%)
Frame = +1
Query: 113 YKILKKTSPQDDSICRTLVFDGGDNDPSPPLPLLPPSISHRRTVFLDLDETLVHS 167
+ +L KT Q+ + LV+ N LPLLPP R L + + +HS
Sbjct: 1297 FYVLVKTWQQNKQLAEWLVYSDSSNKHYRLLPLLPPGHLCSRNNSLLYNPSFLHS 1461
>TC88091 homologue to SP|P51819|HS83_PHANI Heat shock protein 83. [Violet
Japanese morning glory] {Pharbitis nil}, partial (77%)
Length = 1813
Score = 27.7 bits (60), Expect = 6.4
Identities = 31/99 (31%), Positives = 46/99 (46%), Gaps = 8/99 (8%)
Frame = +1
Query: 177 DFVVRPVIDGVPMEFYV----KKRPGVDEFLEALAAK-FEVVVFTAALREYASLVLDRVD 231
D+V R + +G +Y+ KK FLE L K +EV+ A+ EYA L D
Sbjct: 889 DYVTR-MKEGQKDIYYITGESKKAVENSPFLERLKKKGYEVLFMVDAIDEYAVGQLKEYD 1065
Query: 232 RNRLVS---HRLYRDSCKQEDGKLVKDLSLAGRDLKRVV 267
+LVS L D +E+ K ++ + DL RV+
Sbjct: 1066GKKLVSATKEGLKLDDETEEEKKKKEEKKKSFEDLCRVM 1182
>BE320311 similar to PIR|G84781|G847 hypothetical protein At2g36540
[imported] - Arabidopsis thaliana, partial (11%)
Length = 424
Score = 27.7 bits (60), Expect = 6.4
Identities = 29/115 (25%), Positives = 51/115 (44%), Gaps = 20/115 (17%)
Frame = +1
Query: 191 FYVKKRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRV---DRNRLV----SHRLYRD 243
F + KRP +EF++ +FEV V+++A+ L +N+L+ H+
Sbjct: 13 FLLYKRPFSEEFMKFCLERFEVGVWSSAMEHNVDGALACAIGDSKNKLLFVWDQHKCRDS 192
Query: 244 SCKQ-EDGK---LVKDLSLAGRDLKR--------VVIVDDNP-NSFANQPENAIL 285
K E+ K K+L +K+ +++DD P SF N P +I+
Sbjct: 193 GFKSLENNKKPLFFKELKKVWDTIKKGGPYSASNTLLIDDKPYKSFLNPPNTSII 357
>BI308024
Length = 683
Score = 27.7 bits (60), Expect = 6.4
Identities = 12/38 (31%), Positives = 19/38 (49%)
Frame = +1
Query: 147 PPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVI 184
PP + R+ F +T+VH ++ PP S F V+
Sbjct: 196 PPFFINTRSSFFTTGDTVVHYTSSSPPPSSPFTSNTVV 309
>TC82878 similar to PIR|T45677|T45677 ATP-dependent RNA helicase-like
protein - Arabidopsis thaliana, partial (33%)
Length = 1185
Score = 27.3 bits (59), Expect = 8.3
Identities = 10/23 (43%), Positives = 17/23 (73%)
Frame = +3
Query: 51 VKRVPTKSIKDIRRNNNRRWRRK 73
VKR+ T S++ ++R N RW+R+
Sbjct: 453 VKRLKTASLRILKRMNRYRWKRR 521
>AJ504066 weakly similar to PIR|T37811|T378 very hypothetical protein
SPAC17A2.10c - fission yeast (Schizosaccharomyces
pombe), partial (9%)
Length = 580
Score = 27.3 bits (59), Expect = 8.3
Identities = 14/37 (37%), Positives = 23/37 (61%)
Frame = +2
Query: 167 SNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEFL 203
S+T PPP + DF++RP + P + + P VD++L
Sbjct: 170 SSTRPPPSAVDFLLRPPL*LSPSS-CLFRFPAVDQWL 277
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,880,883
Number of Sequences: 36976
Number of extensions: 171805
Number of successful extensions: 1150
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 1120
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1142
length of query: 328
length of database: 9,014,727
effective HSP length: 96
effective length of query: 232
effective length of database: 5,465,031
effective search space: 1267887192
effective search space used: 1267887192
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0118a.3


