

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0114.8
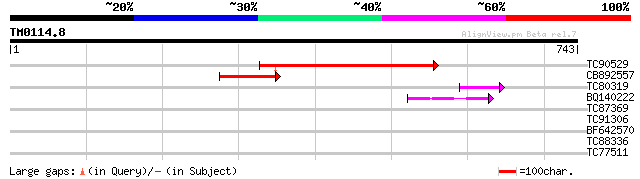
(743 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC90529 similar to GP|18086453|gb|AAL57680.1 At1g28320/F3H9_2 {A... 362 e-100
CB892557 132 5e-31
TC80319 similar to GP|20259261|gb|AAM14366.1 putative HhoA prote... 43 4e-04
BQ140222 homologue to GP|14517500|gb K16N12.18/K16N12.18 {Arabid... 43 5e-04
TC87369 homologue to GP|14517500|gb|AAK62640.1 K16N12.18/K16N12.... 39 0.009
TC91306 weakly similar to PIR|T06371|T06371 probable UDP-glucuro... 32 0.68
BF642570 similar to GP|13172275|gb DegP2 protease {Arabidopsis t... 31 2.0
TC88336 similar to PIR|T51477|T51477 glutamine-rich protein - Ar... 29 5.8
TC77511 similar to GP|14532760|gb|AAK64081.1 putative kinase {Ar... 29 7.5
>TC90529 similar to GP|18086453|gb|AAL57680.1 At1g28320/F3H9_2 {Arabidopsis
thaliana}, partial (6%)
Length = 1017
Score = 362 bits (928), Expect = e-100
Identities = 182/257 (70%), Positives = 201/257 (77%), Gaps = 22/257 (8%)
Frame = +3
Query: 328 QLVIPWEAIVTASSGLLRKW------------------GKGPFVGTYKLD--VPASNTHE 367
QLVIPWEAIV A+SGLLRKW GKGPF+ K + V +SN HE
Sbjct: 195 QLVIPWEAIVNAASGLLRKWPQNTVEGSCYQEGNSCGPGKGPFIDYNKSEAYVLSSNNHE 374
Query: 368 QLNLGSSSLLPIEKAMASVCLVTIGDGVWASGILLNSQGLVLTNAHLLEPWRFGKTHVSG 427
LN G+SS LPIEKAMASVCLVTIGDGVWASGILLNSQGL+LTNAHLLEPWRFGKTH++G
Sbjct: 375 HLNFGNSSPLPIEKAMASVCLVTIGDGVWASGILLNSQGLILTNAHLLEPWRFGKTHITG 554
Query: 428 GG--NGTNSKTLPFMLEGTTSLGNKIESDQISQTLHSKVPVHYPFAADEHGGYKLNPSYD 485
G NGTN + P MLEGTTSL N+ ES Q QTL SK+ YPF A E G YKLN YD
Sbjct: 555 RGYGNGTNPEKFPSMLEGTTSLDNRGESIQTRQTLPSKMTNLYPFVAGEQGRYKLNKPYD 734
Query: 486 NHRNIRIRLDHIKPWVWCDAKVVYICKGPWDVALLQIESIPDNLVPMVMNFSRPSTGSKA 545
+HRNIRIRLDHIKPWVWCDAKVVY+CKGPWDVALLQ+E +PDNL+P++ NFSRPSTGSKA
Sbjct: 735 SHRNIRIRLDHIKPWVWCDAKVVYVCKGPWDVALLQLEPVPDNLLPILTNFSRPSTGSKA 914
Query: 546 YVIGHGLFGPKCGFFPS 562
YVIGHGLFGPKCG P+
Sbjct: 915 YVIGHGLFGPKCGM*PN 965
>CB892557
Length = 300
Score = 132 bits (332), Expect = 5e-31
Identities = 66/81 (81%), Positives = 71/81 (87%), Gaps = 1/81 (1%)
Frame = +1
Query: 276 NSYPPNSSDGSLLM-ADIRTLPGMEGSPVFSEHACLTGVLIRPLRQKTSGVETQLVIPWE 334
N YPPNSSDGSLLM ADIR+LPGMEGSPVFSEHACLTGVLIRPLRQ+TSG E QLVIPWE
Sbjct: 22 NCYPPNSSDGSLLMMADIRSLPGMEGSPVFSEHACLTGVLIRPLRQQTSGAEIQLVIPWE 201
Query: 335 AIVTASSGLLRKWGKGPFVGT 355
AIV A+SGLLRKW + G+
Sbjct: 202 AIVNAASGLLRKWPQNTVEGS 264
>TC80319 similar to GP|20259261|gb|AAM14366.1 putative HhoA protease
precursor {Arabidopsis thaliana}, partial (47%)
Length = 1195
Score = 43.1 bits (100), Expect = 4e-04
Identities = 21/59 (35%), Positives = 34/59 (57%)
Frame = +2
Query: 590 TNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGAVIPHLNFSIPSAAL 648
T G ++T AA++ G SGG +I+S GH++G+ T+ G +NF+IP A+
Sbjct: 803 TEGLSKEAIQTDAAINAGNSGGPLIDSHGHVVGVNTATFTRKGTGASSGVNFAIPIDAV 979
>BQ140222 homologue to GP|14517500|gb K16N12.18/K16N12.18 {Arabidopsis
thaliana}, partial (23%)
Length = 307
Score = 42.7 bits (99), Expect = 5e-04
Identities = 32/114 (28%), Positives = 54/114 (47%), Gaps = 1/114 (0%)
Frame = +1
Query: 522 IESIPDNLVPMVMNFSRPS-TGSKAYVIGHGLFGPKCGFFPSVCSGVIAKVVEAKTTQSY 580
+++ D L P+ + S G K Y IG+ FG V SG+ ++ A T +
Sbjct: 1 VDAPKDKLRPIPVGVSADLLVGXKVYAIGNP-FGLDHTLTTGVISGLRREIXSAATGRPI 177
Query: 581 QSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGA 634
Q + ++T AA++PG SGG +++S G++IG+ T SG +
Sbjct: 178 QDV--------------IQTDAAINPGNSGGPLLDSSGNLIGINTXIYSPSGAS 297
>TC87369 homologue to GP|14517500|gb|AAK62640.1 K16N12.18/K16N12.18
{Arabidopsis thaliana}, partial (42%)
Length = 1210
Score = 38.5 bits (88), Expect = 0.009
Identities = 19/48 (39%), Positives = 33/48 (68%)
Frame = +2
Query: 597 MLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGAVIPHLNFSIP 644
+++T AA++PG SGG +++S G++IG+ T+ SG + + FSIP
Sbjct: 455 VIQTDAAINPGNSGGPLLDSSGNLIGINTAIYSPSGAS--SGVGFSIP 592
>TC91306 weakly similar to PIR|T06371|T06371 probable
UDP-glucuronosyltransferase (EC 2.4.1.-) - garden pea,
partial (31%)
Length = 724
Score = 32.3 bits (72), Expect = 0.68
Identities = 14/29 (48%), Positives = 18/29 (61%)
Frame = +2
Query: 127 RPDLISGVRIDIMTEETNEKSDHGLTPGW 155
RPDL+ G + + +E NE SD GL GW
Sbjct: 59 RPDLVIGGSVVLSSEFVNEISDRGLIAGW 145
>BF642570 similar to GP|13172275|gb DegP2 protease {Arabidopsis thaliana},
partial (29%)
Length = 607
Score = 30.8 bits (68), Expect = 2.0
Identities = 35/135 (25%), Positives = 55/135 (39%), Gaps = 1/135 (0%)
Frame = +2
Query: 516 DVALLQIESIPDNLVPMVMNFSR-PSTGSKAYVIGHGLFGPKCGFFPSVCSGVIAKVVEA 574
D+ALL +ES + F P V+G+ P G SV GV+++V
Sbjct: 155 DLALLSVESEEFWRDVEPLRFGHLPHLQDSVTVVGY----PFGGDTISVTKGVVSRV--- 313
Query: 575 KTTQSYQSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGA 634
SY +E + ++ AA++PG SGG N G IG+ R
Sbjct: 314 -EVTSYAHGSSELLG--------IQIDAAINPGNSGGPAFNDQGECIGVAFXVYRSEDA- 463
Query: 635 VIPHLNFSIPSAALA 649
++ + IPS ++
Sbjct: 464 --ENIGYVIPSTVVS 502
>TC88336 similar to PIR|T51477|T51477 glutamine-rich protein - Arabidopsis
thaliana, partial (67%)
Length = 1297
Score = 29.3 bits (64), Expect = 5.8
Identities = 18/59 (30%), Positives = 30/59 (50%), Gaps = 5/59 (8%)
Frame = +2
Query: 158 AQLLSLFDIPASALGVQS----LIEASLGFSEH-EWEIGWSLASSSNDSKPSRDFLQTQ 211
A+++S I SALG Q ++ +G +H E GW + ++ +PS DFL +
Sbjct: 989 AEMMSRAPIRGSALGSQGSDIGIVGHDMGDQDHGEMMNGWGNNAQRDEKEPSEDFLNDE 1165
>TC77511 similar to GP|14532760|gb|AAK64081.1 putative kinase {Arabidopsis
thaliana}, partial (58%)
Length = 1348
Score = 28.9 bits (63), Expect = 7.5
Identities = 36/140 (25%), Positives = 59/140 (41%), Gaps = 5/140 (3%)
Frame = +3
Query: 578 QSYQSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGAVIP 637
QSY M G A L + + PGAS I +IG T+++ G +
Sbjct: 393 QSYIYSSKVDMWAMGAIMAELFSLRPLFPGASEADEIYKICGVIGNPTTDSWADGLKLAR 572
Query: 638 HLNFSIPSAA---LAPIFKFAEDMKDLSLLQILD--EPNEYLSSIWALMRPSYPKLHSVP 692
+N+ P A L+ + A D +SL+Q L +P + ++ AL P + +P
Sbjct: 573 DINYQFPQLAGVNLSALIPSASD-HAISLIQSLCSWDPCKRPTASEALQHPFFQSCFYIP 749
Query: 693 GSPQSLVDKKSKEEKGSRFA 712
S +S ++ G+R A
Sbjct: 750 PSLRSRAVARTPPPAGTRGA 809
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.135 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,260,718
Number of Sequences: 36976
Number of extensions: 342247
Number of successful extensions: 1529
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 1520
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1526
length of query: 743
length of database: 9,014,727
effective HSP length: 103
effective length of query: 640
effective length of database: 5,206,199
effective search space: 3331967360
effective search space used: 3331967360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0114.8


