

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0114.7
(1898 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
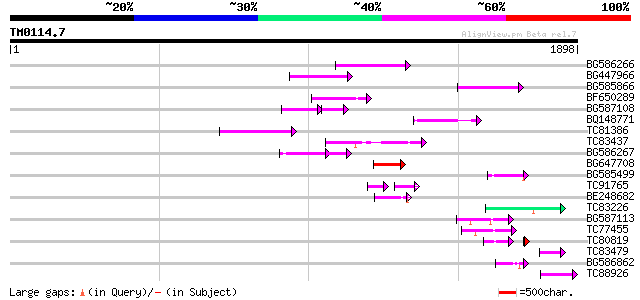
Score E
Sequences producing significant alignments: (bits) Value
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 193 5e-49
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 151 3e-36
BG585866 146 7e-35
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 131 2e-30
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 70 8e-25
BQ148771 109 1e-23
TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR re... 105 2e-22
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 102 1e-21
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 93 1e-18
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 82 2e-15
BG585499 75 2e-13
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 55 3e-13
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 67 5e-11
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 66 1e-10
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 61 4e-09
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 56 2e-07
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 44 7e-06
TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protei... 50 1e-05
BG586862 48 4e-05
TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {D... 46 1e-04
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 193 bits (491), Expect = 5e-49
Identities = 101/253 (39%), Positives = 152/253 (59%), Gaps = 1/253 (0%)
Frame = -3
Query: 1091 VRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHM 1150
V + R I+ CN +K+I K+L R++P LR +I P QS+F+PGR DN + +++H++
Sbjct: 778 VSEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYL 599
Query: 1151 SKSTAKRGM-VAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILWN 1209
+S AK+ + +A K D+ KAYD ++W+FL+E L GF + I+ IM V++ S L N
Sbjct: 598 RQSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLIN 419
Query: 1210 GCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHLF 1269
G P RGLRQGDP+SPYLF+LC E LS L Q + KG +++++N PPI+HL
Sbjct: 418 GGPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARNCPPINHLL 239
Query: 1270 FADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVAP 1329
FADD + F +++ + +L S M + +SG IN +KS S S+ + D +
Sbjct: 238 FADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAIIDRVKGELK 59
Query: 1330 IPFVHDLGKYLGF 1342
I GKYLG+
Sbjct: 58 IAKEGGTGKYLGY 20
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 151 bits (381), Expect = 3e-36
Identities = 82/209 (39%), Positives = 116/209 (55%)
Frame = +2
Query: 938 GDRNTAYFHTQTLIRRRKNRVHRLKLEDGTWCGDEEVLKRVVHGFFVNLFTGITPLASPR 997
GD+NT +FH++ RR+ N + +LK E G WC EE ++R++ +F NLFT P A
Sbjct: 17 GDKNTKFFHSKASQRRKVNEIKKLKDETGNWCKGEENVERLLITYFNNLFTSSNPTAIEE 196
Query: 998 VTHDLFPSLSVEAKSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSL 1057
+ LS E + ++EV A+ M+ APG DG F++KYW+ VG +
Sbjct: 197 TCEVVKGKLSHEHIVWCEKEFTEEEVLEAINQMHPVKAPGPDGLPALFFQKYWHIVGKEV 376
Query: 1058 WHMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLRP 1117
MV + E L +VLIPK P+T +D RPISLCNV K+ITKV+ NR++
Sbjct: 377 QQMVLQVLNNSMETEELNKTFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVIANRVKQ 556
Query: 1118 FLRKLIGPMQSSFLPGRGTMDNAFLAQEV 1146
L +I QS+F+ GR DNA +A V
Sbjct: 557 TLPDVIDVEQSAFVQGRLITDNALIAWSV 643
>BG585866
Length = 828
Score = 146 bits (369), Expect = 7e-35
Identities = 68/222 (30%), Positives = 113/222 (50%), Gaps = 1/222 (0%)
Frame = +3
Query: 1500 LLKARDRLHTGFVFRLGNGETSLWHDDWSGMGNIAPAVPYVDIHDIDRRLCDLVDNGTWN 1559
+++ ++ L +G+ +R G+G +S W+ +WS +G + P+VDIHD+ + D+ G +
Sbjct: 84 IIRDKNVLKSGYTWRAGSGNSSFWYTNWSSLGLLGTQAPFVDIHDLHLTVKDVFTTGGQH 263
Query: 1560 MQVLYTSLPVDVLERLQKIKPTIVPNRQDVWTWETNSMGIYTVRGAYRWLQDQQSHLPVV 1619
Q LYT LP D+ E + + D + W NS G+YT + Y W+ Q +
Sbjct: 264 TQSLYTILPTDIAEVINNTHLNFNASIGDAYIWPHNSNGVYTAKSGYSWILSQTETVNYN 443
Query: 1620 D-DWNWIWKLKVPEKIRTFVWLTLQNSLQVNLHRFRCKMAASPSCSRCSAPEEDILHCLR 1678
+ W+WIW+LK+PEK + F+WL N++ M S CSRC EE HC+R
Sbjct: 444 NSSWSWIWRLKIPEKYKFFLWLACHNAVPTLSLLNHRNMVNSAICSRCGEHEESFFHCVR 623
Query: 1679 TCPHSQELWLKFGAFAWPNFTAGDHSSWIRSQARSNNGVKFI 1720
C S+ +W K G + F++ W++ + F+
Sbjct: 624 DCRFSKIIWHKIGFSSPDFFSSSSVQDWLKDGISCHRPTTFL 749
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 131 bits (330), Expect = 2e-30
Identities = 72/201 (35%), Positives = 118/201 (57%), Gaps = 1/201 (0%)
Frame = +3
Query: 1011 KSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKV 1070
+ LL EV++AL SM+S APG DG+ F+K WN +GDS+ + + F+ G +
Sbjct: 3 QDLLCSEFTAVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFM 182
Query: 1071 NENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSF 1130
+ + V L+PK ++V++ RPI+ C+V +K+I+K+L +R++ L ++ QS+F
Sbjct: 183 PKIINCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAF 362
Query: 1131 LPGRGTMDNAFLAQEVIHHMS-KSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPE 1189
+ GR DN L+ E++ S K + R MV KIDL KAYDS W F++ + GFP
Sbjct: 363 VKGRVIFDNIILSHELVKSYSRKGISPRCMV--KIDLXKAYDSXEWPFIKHLMLELGFPY 536
Query: 1190 VTINLIMSSVTSSQVSILWNG 1210
+N +M+ +T++ + NG
Sbjct: 537 KFVNWVMAXLTTASYTFNXNG 599
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 69.7 bits (169), Expect(2) = 8e-25
Identities = 36/89 (40%), Positives = 53/89 (59%)
Frame = +3
Query: 1045 FYKKYWNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAF 1104
F++ W+ + L MV GK++ L + LIPK +P+ + +LRPISLCNV +
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 1105 KVITKVLVNRLRPFLRKLIGPMQSSFLPG 1133
K+I+KVL RL+ L LI QS+F+ G
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFVHG 677
Score = 64.3 bits (155), Expect(2) = 8e-25
Identities = 41/137 (29%), Positives = 68/137 (48%), Gaps = 1/137 (0%)
Frame = +2
Query: 911 LQREYRNILRQEELL*YQKARENRVHL-GDRNTAYFHTQTLIRRRKNRVHRLKLEDGTWC 969
L+ + + + EE QK+R N H+ GD N ++H T R +NR+ L DG W
Sbjct: 8 LKEKLQEAYKDEEDYWQQKSR-NMWHISGDLNKKFYHALTKQRHARNRIVGLYDYDGNWI 184
Query: 970 GDEEVLKRVVHGFFVNLFTGITPLASPRVTHDLFPSLSVEAKSLLLQPVRKDEVRHALMS 1029
+E+ +++V +F +LF TP ++ S++ + LL+ ++EVR AL
Sbjct: 185 TEEQGVEKVAVDYFEDLFQRTTPTGFDGFLDEITSSITPQMNQRLLRLATEEEVRLALFI 364
Query: 1030 MNSYTAPGADGFQPFFY 1046
M+ APG DG +
Sbjct: 365 MHPEKAPGPDGMTTLLF 415
>BQ148771
Length = 680
Score = 109 bits (272), Expect = 1e-23
Identities = 74/229 (32%), Positives = 108/229 (46%), Gaps = 1/229 (0%)
Frame = -3
Query: 1351 RNRFNFLLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQVFNMPRGVTCKINQL 1410
++RF+ + + L +W+AN LSLA RV LAKSVI A+P Y M +P+ +I +L
Sbjct: 612 KSRFSVYYQ-VHVMLANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQKL 436
Query: 1411 IRSFIWSGKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIWCLMHKPNKLW 1470
R F+W +H V WE +++PK GLG+R ++ N A + K W + N L
Sbjct: 435 QRKFVWGDTEVSRRYHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSNSLC 256
Query: 1471 VRVLSHKYLSNSSVLQV-QAKPQDSQVWKGLLKARDRLHTGFVFRLGNGETSLWHDDWSG 1529
V+ KY + S+ ++ KP DS +WK L+K LW
Sbjct: 255 TEVMRGKYQRSESLEEIFLEKPTDSSLWKALVK-------------------LW------ 151
Query: 1530 MGNIAPAVPYVDIHDIDRRLCDLVDNGTWNMQVLYTSLPVDVLERLQKI 1578
+I+R L D NG WN + L LP DVL+R+ I
Sbjct: 150 -------------PEIERNLVD--SNGNWNWEKLKQWLPFDVLQRIMAI 49
>TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 798
Score = 105 bits (261), Expect = 2e-22
Identities = 76/266 (28%), Positives = 122/266 (45%), Gaps = 8/266 (3%)
Frame = -1
Query: 701 REILWRHMTLLRRRFLLPWLLLGDFNEILFPSEVRGGDFLPNRAAM--FASVLDTCQLVD 758
R LW ++ ++ + L W +GDFN IL E +G P R M F D L
Sbjct: 795 RRQLWSAISNIQTQHKLSWCCIGDFNTILGSHEHQGSH-TPARLPMLDFQQWSDVNNLFH 619
Query: 759 LGAVGRRFTWFQKANDRLILSKRLDRALGDMEWKVAFPDAVVDVLNRVHSDHCPILLQCG 818
L G FTW R KRLDR++ + L ++ SDH PIL +
Sbjct: 618 LPTRGSAFTWTNGRRGRNNTRKRLDRSIVNQLMIDNCESLSACTLTKLRSDHFPILFELQ 439
Query: 819 SPQTLAHNRPFRFLAAWADHPDFAAVVDNAWRSGEE-----CITSKLERVREASTVFNKE 873
+ Q + + F+F+ W+ HPD ++ W + + KL+ ++E V+NK
Sbjct: 438 T-QNIQFSSSFKFMKMWSAHPDCINIIKQCWANQVVGCPMFVLNQKLKNLKEVLKVWNKN 262
Query: 874 VFGNIFRRKRHVEARLRGVQRELDRRVTSDMVLF-EAELQREYRNILRQEELL*YQKARE 932
FGN+ + + L +Q ++D SD+++ E Q + L EE+ ++K++
Sbjct: 261 TFGNVHSQVDNAYKELDDIQVKIDSIGYSDVLMDQEKAAQLNLESALNIEEVFWHEKSKV 82
Query: 933 NRVHLGDRNTAYFHTQTLIRRRKNRV 958
N GDRNTA+FH I+R + +
Sbjct: 81 NWHCEGDRNTAFFHRVAKIKRTSSLI 4
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 102 bits (254), Expect = 1e-21
Identities = 92/351 (26%), Positives = 152/351 (43%), Gaps = 14/351 (3%)
Frame = +2
Query: 1058 WHMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLRP 1117
+ V E + K+ + + + LIPKVD P + D RPISL +K++ K+L NRLR
Sbjct: 2 FRFVSEFHRNRKLFKGINSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRV 181
Query: 1118 FLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHMSKSTAK---------RGMVAFKIDLEK 1168
+ +I QS+F+ R ++ FL Q + ++ K + ++ I L
Sbjct: 182 VIGSVISDAQSAFVKNRQILEMVFL*QMRLWMRLRN*RKIFCCLRWILKRLITLSIGLIW 361
Query: 1169 AYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPM 1228
V SFL + I V+++ S+L NG
Sbjct: 362 ILF*VGMSFLV----------LWRKWIKECVSTATTSVLVNG------------------ 457
Query: 1229 SPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPI-SHLFFADDVLLFCEASVAQVNL 1287
SP +VL++SLV + P + SHL FA+D LL + A +
Sbjct: 458 SP---------TNVLMKSLVQTQLFTRYSFGVVNPVVVSHLQFANDTLLLETKNWANIRA 610
Query: 1288 LASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISV----VAPIPFVHDLGKYLGFP 1343
L + + +F SGLK+N KS + + ++ SV V +PF+ YLG P
Sbjct: 611 LRAALVIF*AMSGLKVNFHKSGLVCVNIAPSWLSEAASVLSWKVGKVPFL-----YLGMP 775
Query: 1344 LKGGRIHRNRFNFLLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTM 1394
++G + + ++ I+ +L W + LS GR+ L KSV+ ++ Y +
Sbjct: 776 IEGNSRRLSFWEPIVNRIKARLTGWNSRFLSFGGRLVLLKSVLTSLSVYAL 928
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 92.8 bits (229), Expect = 1e-18
Identities = 54/172 (31%), Positives = 90/172 (51%)
Frame = +3
Query: 903 DMVLFEAELQREYRNILRQEELL*YQKARENRVHLGDRNTAYFHTQTLIRRRKNRVHRLK 962
++ L +EL EY N EE+ QK+R N + GDRNT +FH T RR +NR+ L
Sbjct: 78 ELSLLRSELNEEYHN----EEIFWMQKSRLNWLRSGDRNTKFFHAVTKNRRAQNRILSLI 245
Query: 963 LEDGTWCGDEEVLKRVVHGFFVNLFTGITPLASPRVTHDLFPSLSVEAKSLLLQPVRKDE 1022
+D EE L R+ F L++ + + + ++ E + L+ + ++E
Sbjct: 246 DDDDKEWFVEEDLGRLADSHFKLLYSSEDVGITLEDWNSIPAIVTEEQNAQLMAQISREE 425
Query: 1023 VRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNENL 1074
VR A+ +N + PG DG FF++++W+ +GD L M +E + GK+ E +
Sbjct: 426 VREAVFDINPHKCPGPDGMNVFFFQQFWDTMGDDLTSMAQEFLRTGKLEEGI 581
Score = 50.1 bits (118), Expect = 8e-06
Identities = 29/91 (31%), Positives = 48/91 (51%), Gaps = 5/91 (5%)
Frame = +1
Query: 1058 WHMV-----KEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLV 1112
W M+ K + ++G + L+PK + + + RPISLCNVA+K+++KVL
Sbjct: 517 WEMISHLWRKNSSEQGNLKRESTKQTSGLVPKKLEAKRLVEFRPISLCNVAYKIVSKVLS 696
Query: 1113 NRLRPFLRKLIGPMQSSFLPGRGTMDNAFLA 1143
RL+ L +I Q++F + DN +A
Sbjct: 697 KRLKSVLPWIITETQAAFGRRQLISDNILIA 789
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 82.4 bits (202), Expect = 2e-15
Identities = 41/107 (38%), Positives = 65/107 (60%)
Frame = +1
Query: 1218 PGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLF 1277
P +GLRQGDP+SPYLF+LC LS L++ +K + I+++++ P I+HL FADD LLF
Sbjct: 4 PEKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLF 183
Query: 1278 CEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSI 1324
A++ + + + + +SG +N KS+ S+ V K+ I
Sbjct: 184 ARANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQNVPNQEKEMI 324
>BG585499
Length = 792
Score = 75.5 bits (184), Expect = 2e-13
Identities = 41/149 (27%), Positives = 70/149 (46%), Gaps = 11/149 (7%)
Frame = +3
Query: 1600 YTVRGAYRWLQDQQSHLPVVD-DWNWIWKLKVPEKIRTFVWLTLQNSLQVNLHRFRCKMA 1658
+ ++ +Y L QS +VD DW +W + P + +TF+WL + N R R
Sbjct: 165 FKIKSSYNLLV*DQS---IVDCDWKMLWGWRGPHRTQTFMWLVAHGCILTNYRRSRWGTR 335
Query: 1659 ASPSCSRCSAPEEDILHCLRTCPHSQELWLKFGAFAWPN--FTAGDHSSWI-RSQARSNN 1715
+C C +E +LH L C + ++W++ W F+ D W+ ++ ++ +N
Sbjct: 336 VLATCPCCGNADETVLHVLCDCRPASQVWIRLVPSDWITNFFSFDDCRDWVFKNLSKRSN 515
Query: 1716 GVK-------FIVGLWGVWKWRNNMIFED 1737
GV F+ W +W WRN IFE+
Sbjct: 516 GVSKFKWQPTFMTTCWHMWTWRNKAIFEE 602
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 55.1 bits (131), Expect(2) = 3e-13
Identities = 26/68 (38%), Positives = 40/68 (58%)
Frame = +2
Query: 1199 VTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIRL 1258
V S+ +L N + P RGL+QGD +SPY+F++C+E LS LI ++GD +
Sbjct: 107 VESNDYYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERGDTHGTSI 286
Query: 1259 SKNGPPIS 1266
+ PP+S
Sbjct: 287 *RGAPPVS 310
Score = 40.0 bits (92), Expect(2) = 3e-13
Identities = 25/85 (29%), Positives = 43/85 (50%)
Frame = +3
Query: 1287 LLASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFPLKG 1346
++ + + L+ + SG I+L KS S+ V +++K SI+ + + F+ KYLG P
Sbjct: 336 IMKNILILYEEDSGKAISLRKS*IYCSRNVPDILKTSITYILGVQFMLGTCKYLGLPSMI 515
Query: 1347 GRIHRNRFNFLLESIQRKLGSWRAN 1371
GR F+ + K G W+ N
Sbjct: 516 GRDRTTTFSSI------KGGVWQKN 572
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 67.4 bits (163), Expect = 5e-11
Identities = 45/132 (34%), Positives = 73/132 (55%), Gaps = 8/132 (6%)
Frame = +3
Query: 1220 RGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLFCE 1279
RGL+QGDP++P+LF+L E +S L+++ V++ ++ + + G +SHL +ADD L
Sbjct: 24 RGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTRVSHLQYADDTLCIGM 203
Query: 1280 ASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVA--------PIP 1331
+V + L + +Q F +SGLK+N KS I G++ V +D + IP
Sbjct: 204 PTVDNLWTLKALLQGFEMASGLKVNFHKSSLI---GIN-VPRDFMEAACRFLNCREESIP 371
Query: 1332 FVHDLGKYLGFP 1343
F+ YLG P
Sbjct: 372 FI-----YLGLP 392
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 65.9 bits (159), Expect = 1e-10
Identities = 68/293 (23%), Positives = 114/293 (38%), Gaps = 26/293 (8%)
Frame = +3
Query: 1592 WETNSMGIYTVRGAYR----WLQDQQSHLPVVDD----WNWIWKLKVPEKIRTFVWLTLQ 1643
W N GIY+V+ Y W Q ++ D W IW L + + +W L
Sbjct: 6 WMHNPTGIYSVKSGYNTLRTWQTQQINNTSTSSDETLIWKKIWSLHTIPRHKVLLWRILN 185
Query: 1644 NSLQVNLHRFRCKMAASPSCSRCSAPEEDILHCLRTCPHSQELW------LKFGAFAWPN 1697
+SL V + + P C RC + E I H +CP S+ +W + F PN
Sbjct: 186 DSLPVRSSLRKRGIQCYPLCPRCHSKTETITHLFMSCPLSKRVWFGSNLCINFDNLPNPN 365
Query: 1698 FTAGDHSSWIRSQARSNNGVKFIVGLWGVWKWRNNMIFEDSPWSLEEAWRRVCH------ 1751
F + + + + I+ + +W RN + ED + +R +
Sbjct: 366 FIN*LYEAIL*KDECITI*IAAII--YNLWHARNLSVLEDQTILEMDIIQRASNCISDYK 539
Query: 1752 ----EHDEIVAVLGEEAGSMD-CWLGSRWQPPMAGSIKLNVDGSYRDVDDSSGVGGLARD 1806
+ +A G + S ++W+ P G +K+N D + ++ G+G + RD
Sbjct: 540 QANTQAPPSMARTGYDPRSQHRPAKNTKWKRPNLGLVKVNTDANLQN-HGKWGLGIIIRD 716
Query: 1807 PSGNWLFGFLAHRRGGN-AFLAEAQALLLGLELVWARGYRDIVVEVDCADLLQ 1858
G + G + A AEA ALL G+ G+ + E D L++
Sbjct: 717 EVGLVMAASTWETDGNDRALEAEAYALLTGMRFAKDCGFXKVXFEGDNEKLMK 875
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 61.2 bits (147), Expect = 4e-09
Identities = 50/217 (23%), Positives = 87/217 (40%), Gaps = 23/217 (10%)
Frame = -3
Query: 1494 SQVWKGLLKARDRLHTGFVFRLGNGE-TSLWHDDWSGMGNIAPAVPYVD---------IH 1543
S W+ + A+ + G +GNGE T++W + + P ++
Sbjct: 735 SYAWRSIHSAQHLIKQGAKVIIGNGENTNIWEREMAWKLTCVTNHPNKHSSRAY*APTLY 556
Query: 1544 DIDRRLCDLVDNGTWNMQVLYTSLPVDVLERLQKIKPTIVPNRQDVWTWETNSMGIYTVR 1603
+ D N ++ + P ++ I P P +D ++WE + G Y+V+
Sbjct: 555 GYEGCRSDDPMRRERNANLINSIFPEGTRRKILSIHPQ-GPIGEDSYSWEYSKSGHYSVK 379
Query: 1604 GAY------------RWLQDQQSHLPVVDD-WNWIWKLKVPEKIRTFVWLTLQNSLQVNL 1650
Y R DQ P +DD + +WK K+R F+W + NSL
Sbjct: 378 SGYYVQTNIIAAANQRGTVDQ----PSLDDLYQRVWKYNTSPKVRHFLWRCISNSLPTAA 211
Query: 1651 HRFRCKMAASPSCSRCSAPEEDILHCLRTCPHSQELW 1687
+ ++ SCSRC E + H L CP+++ +W
Sbjct: 210 NMRSRHISKDGSCSRCGMESETVNHILFQCPYARLIW 100
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 55.8 bits (133), Expect = 2e-07
Identities = 48/210 (22%), Positives = 86/210 (40%), Gaps = 26/210 (12%)
Frame = -2
Query: 1514 RLGNGETSL-WHDDWSGMGNIAPAVP--YVDIHDIDRRLCDLVDNGT--------WNMQV 1562
++G+G +S W D W + + P + + DL D W
Sbjct: 703 KVGDGRSSFFWKDAWDSSVPLRESFPRAFFPYRLLKMGCGDLWDMNAEGVRWRLYWRRLE 524
Query: 1563 LYTSLPVDVLERLQKIKPTIVPNRQDVWTWETNSMGIYTVRGAY------RWLQDQQSHL 1616
L+ +LE L +++ ++ D+W W+ + G+++V Y R L+D+ S+
Sbjct: 523 LFEWEKERLLELLGRLEGVVLRYWADIWVWKPDKEGVFSVNSCYFLLQNLRLLEDRLSYE 344
Query: 1617 PVVDDWNWIWKLKVPEKIRTFVWLTLQNSL--QVNLHRFR-CKMAASPSCSRCSAPEEDI 1673
V + +WK K P K+ F W + + VNL + R ++ S C C +E +
Sbjct: 343 EEVI-FRELWKSKAPAKVLAFSWTLFLDRIPTMVNLGKRRLLRVEDSKRCVFCGCQDETV 167
Query: 1674 LHCLRTCPHSQEL------WLKFGAFAWPN 1697
+H C ++ WL F + PN
Sbjct: 166 VHLFLHCDVISKV*REVMRWLNFNLISPPN 77
Score = 42.4 bits (98), Expect = 0.002
Identities = 21/54 (38%), Positives = 28/54 (50%)
Frame = -3
Query: 1425 WHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIWCLMHKPNKLWVRVLSHKY 1478
WH + P+ +GGLGVRD L N +L+ K W L+ + LW VL Y
Sbjct: 975 WHCL-------PRCKGGLGVRDIRLVNVSLLAKWWWRLLQDQSSLWKEVLEDIY 835
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 43.5 bits (101), Expect(2) = 7e-06
Identities = 30/107 (28%), Positives = 44/107 (41%), Gaps = 4/107 (3%)
Frame = +2
Query: 1585 NRQDVWTWETNSMGIYTVRGAYRWLQDQQ--SHLPVVDDWNWIWKLKVPEKIRTFVWLTL 1642
N D W W + + Y+V+ YR++ S +VDD +W +P K+ FVW L
Sbjct: 509 NVNDKWRWLLDPVNGYSVKVFYRYITSTGHISDRSLVDD---VWHKHIPSKVSLFVWRLL 679
Query: 1643 QNSLQV--NLHRFRCKMAASPSCSRCSAPEEDILHCLRTCPHSQELW 1687
+N L NL +A + +C E H C LW
Sbjct: 680 RNRLPTKDNLVHRGVLLATNAACVCGCVDSESTTHLFLHCNVFCSLW 820
Score = 26.2 bits (56), Expect(2) = 7e-06
Identities = 8/18 (44%), Positives = 13/18 (71%)
Frame = +1
Query: 1721 VGLWGVWKWRNNMIFEDS 1738
V W +WK RNN +F+++
Sbjct: 937 VNFWVIWKERNNRLFQNT 990
>TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protein {Plasmodium
falciparum 3D7}, partial (0%)
Length = 1222
Score = 49.7 bits (117), Expect = 1e-05
Identities = 30/84 (35%), Positives = 43/84 (50%)
Frame = +2
Query: 1775 WQPPMAGSIKLNVDGSYRDVDDSSGVGGLARDPSGNWLFGFLAHRRGGNAFLAEAQALLL 1834
W+ P G KLN DGS +++G GGL RD G + F++ G+ FL E A+
Sbjct: 977 WKKPEIGWTKLNTDGSVN--KETAGFGGLLRDYRGEPICAFVSKAPQGDTFLVELWAIWR 1150
Query: 1835 GLELVWARGYRDIVVEVDCADLLQ 1858
GL L G + I VE D +++
Sbjct: 1151 GLVLSLGLGIKSIWVESDSMSVVK 1222
>BG586862
Length = 804
Score = 47.8 bits (112), Expect = 4e-05
Identities = 33/123 (26%), Positives = 54/123 (43%), Gaps = 10/123 (8%)
Frame = -1
Query: 1625 IWKLKVPEKIRTFVWLTLQNSLQVNLHRFRCKMAASPSCSRCSAPEEDILHCLRTCPHSQ 1684
+W +K + ++F+W L N+L V + + S C RC + E + H C +Q
Sbjct: 654 VWGIKTIPRHKSFLWRLLHNALPVKDELHKRGIRCSLLCPRCESKIETVQHLFLNCEVTQ 475
Query: 1685 ELWLKFGAFAWPNFTAGDHSS-------WIRSQARSNNG---VKFIVGLWGVWKWRNNMI 1734
+ W FG+ NF HSS WI + N+ + L+ +W RN +
Sbjct: 474 KEW--FGSQLGINF----HSSGVLHFHDWITNFILKNDEETIIALTALLYSIWHARNQKV 313
Query: 1735 FED 1737
FE+
Sbjct: 312 FEN 304
>TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {Drosophila
melanogaster}, partial (1%)
Length = 1073
Score = 46.2 bits (108), Expect = 1e-04
Identities = 43/129 (33%), Positives = 65/129 (50%), Gaps = 6/129 (4%)
Frame = +3
Query: 1776 QPPMAGSIKLNVDGSY---RDVDDSSGVGGLARDPSGNWLFGFLAHRRGGNAFL--AEAQ 1830
+P + LNVDGS R+V S+G GG+ D SG WL GF A + N + E +
Sbjct: 321 RPDYLEEVILNVDGSLLREREVP-SAGCGGVLSDSSGKWLCGF-AQKLNPNLKVDETEKE 494
Query: 1831 ALLLGLELVWARGYRDIVVEVDCADLLQSLDDEDRRRFLPILGDIRN-MKDRGWRISLER 1889
A+L GL V +G R I+V+ D ++ S++ R P++ IR+ + W +L
Sbjct: 495 AILRGLLWVKEKGKRKILVKSDNEGVVYSVNCGGRSND-PLVCGIRDLLNSPHWEATLTC 671
Query: 1890 VRRDCNAPA 1898
+ NA A
Sbjct: 672 IHGRSNAVA 698
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.332 0.143 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 68,830,669
Number of Sequences: 36976
Number of extensions: 1133249
Number of successful extensions: 6544
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 4719
Number of HSP's successfully gapped in prelim test: 207
Number of HSP's that attempted gapping in prelim test: 1575
Number of HSP's gapped (non-prelim): 5246
length of query: 1898
length of database: 9,014,727
effective HSP length: 110
effective length of query: 1788
effective length of database: 4,947,367
effective search space: 8845892196
effective search space used: 8845892196
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0114.7


