

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
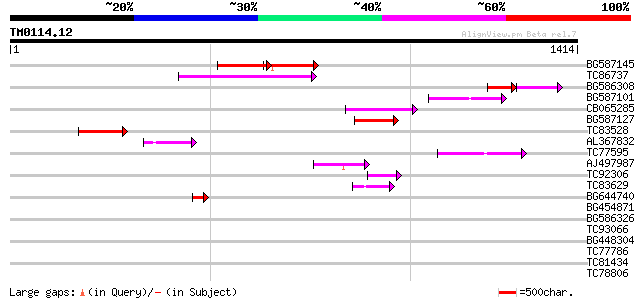
Query= TM0114.12
(1414 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 175 1e-43
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 154 2e-37
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 83 1e-25
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 114 2e-25
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 107 4e-23
BG587127 weakly similar to PIR|H84506|H84 probable retroelement ... 97 4e-20
TC83528 92 1e-18
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 82 2e-15
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 77 6e-14
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 65 1e-10
TC92306 53 7e-07
TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNa... 52 2e-06
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 48 3e-05
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 40 0.006
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 39 0.019
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 37 0.042
BG448304 37 0.042
TC77786 37 0.054
TC81434 28 0.40
TC78806 similar to GP|8978273|dbj|BAA98164.1 receptor protein ki... 34 0.46
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 175 bits (443), Expect = 1e-43
Identities = 81/136 (59%), Positives = 107/136 (78%)
Frame = +2
Query: 518 MHPSDEESTAFMTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVK 577
MHP D E TAF+T++ YCYK MPFGLKNAG+TYQRL++++F+ ++G MEVY+DDM+VK
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 578 SARASDHGGDLKEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAIL 637
S RA+DH LKE F L Y MKLNP KC+FG+ G+FLG+++T +GIEVNP + AIL
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 638 EMKSPTSVKEVQRLTG 653
++ SP + +EVQRLTG
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 112 bits (280), Expect = 1e-24
Identities = 61/146 (41%), Positives = 89/146 (60%), Gaps = 7/146 (4%)
Frame = +3
Query: 632 KGRAILEMKSPTSVKEVQRLT-------GRMAALSRFLPMAGDKAAPFFTCLKKNSKFQW 684
+ R IL P VQR+ GR+AAL+RF+ + DK PF+ L N +F W
Sbjct: 324 ESR*ILSRSPPY*TSLVQRIAERSSDSRGRIAALNRFISRSTDKCLPFYKLLCGNKRFVW 503
Query: 685 TEECEQAFTKLKETLATLPVLSKPTPGVPLVLYLAVTDKAVSTVLLQEEGKKQKVIYFVS 744
E+CE+AF +LK+ L T PVLSKP G L LY+A++ AVS+VL++E+ +QK I++ S
Sbjct: 504 DEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIAISSTAVSSVLIREDRGEQKPIFYTS 683
Query: 745 HTLQGAELRYQKIEKAALAILKTARR 770
+ E RY +EK A A++ +AR+
Sbjct: 684 KRMTDPETRYPTLEKMAFAVITSARK 761
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 154 bits (390), Expect = 2e-37
Identities = 102/347 (29%), Positives = 167/347 (47%), Gaps = 3/347 (0%)
Frame = +1
Query: 421 PMRRMSEEKHKAVQLETEKLIKARFIREVQYPTWLANVVMVKKANGKWRMCTDYTSLNKV 480
P+ MS ++ ++ E L+ FI+ A V+ V+K G R C DY +LN +
Sbjct: 475 PLYGMSRQELLVLKKTLEDLLDKGFIKASGSAAG-APVLFVRKPGGGIRFCVDYRALNAI 651
Query: 481 CPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAFMTNQANYCYKTM 540
KD YPLP + + + +G + +D + ++++ + D+E TAF T + +
Sbjct: 652 TKKDRYPLPLISETLRRVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVC 831
Query: 541 PFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIV-KSARASDHGGDLKEAFDQLRTYQ 599
PFGL A AT+QR ++K + + + Y+DD+++ + DH ++ +L
Sbjct: 832 PFGLTGAPATFQRYINKTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAG 1011
Query: 600 MKLNPEKCSFGIQGGKFLGFMLTS-RGIEVNPDKGRAILEMKSPTSVKEVQRLTGRMAAL 658
+ L+P+KC F + K++GF+LT+ +G+ +P K AI + P SVK + G
Sbjct: 1012LSLDPKKCEFSVTTVKYVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYY 1191
Query: 659 SRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLSKPTPGVPLVLYL 718
F+P + P +K+ F+W E E AFTKLK A PVL P +
Sbjct: 1192KDFIPGYSEITEPLTRLTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVET 1371
Query: 719 AVTDKAVSTVLLQEEGK-KQKVIYFVSHTLQGAELRYQKIEKAALAI 764
+ A+ VL QE+G + F S L AE Y +K LA+
Sbjct: 1372DCSGFALGGVLTQEDGTGAAHPVAFHSQRLSPAEYNYPIHDKELLAV 1512
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 83.2 bits (204), Expect(2) = 1e-25
Identities = 38/72 (52%), Positives = 51/72 (70%)
Frame = -2
Query: 1193 GVPMALVMDNGTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESANKVILKGLKKKLD 1252
G+P +V DNG+ F S+ REFC I + AS +PQ+NGQAE++NK+I+ GLKK+LD
Sbjct: 682 GLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRLD 503
Query: 1253 EAKGLWAEELPG 1264
KG WA+EL G
Sbjct: 502 LKKGCWADELDG 467
Score = 53.1 bits (126), Expect(2) = 1e-25
Identities = 34/120 (28%), Positives = 61/120 (50%), Gaps = 5/120 (4%)
Frame = -3
Query: 1264 GVLWAYNTTEQSSTKETPYRLTYGTDAMLSVEIENQSWRVARFNEND--NGENLIANLIM 1321
GVLW++ T + +TK TP+ + + +AM E+ S +R +N N + L L
Sbjct: 468 GVLWSHRTNPRGATKSTPFSMAHRVEAMAPAEVNVTSL*RSRMPQNIELNNDRLFNALET 289
Query: 1322 LPEEQREAHIRNEAGKVKVARKFSTKVVPRKMRVGDLVL*K---NTIPDKHNKLSPNWGG 1378
+ E + +A +R + + ++ ++ V + +++GD+VL K NT KL NW G
Sbjct: 288 IEERRDQALLRIQNYQHQIESYYNKTVKSQPLKLGDIVLCKVFENTKELNAGKLGTNWEG 109
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 114 bits (286), Expect = 2e-25
Identities = 62/194 (31%), Positives = 99/194 (50%)
Frame = +2
Query: 1045 LYRRGIMSPMLKCVDTKDALGIMAEVHEGVCSSHIGGRSLAVKVIRAGFYWPTMKKDCLE 1104
LY++ + ++CV ++ GI+ H + H K+ +AGF+WPTM KD
Sbjct: 41 LYKQCADNIYIRCVAEEEIPGILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHS 220
Query: 1105 YVKKCEKCQVFSDLHKAPPEELTTMMAPWPFAMWGTDILGPFPVAKAQMKYIIVAVDYFT 1164
++ KC+ CQ ++ ++ F +WG D +GPFP + KYI+VAVDY +
Sbjct: 221 FISKCDPCQRQGNIS*RNEMPQNFILEVEVFDVWGIDFMGPFP-SSYNNKYILVAVDYVS 397
Query: 1165 KWIEAEAVATITAAKVRNFLWQRIVCRFGVPMALVMDNGTQFTSSVTREFCAEMGIEMRF 1224
KW+EA A T A V I RFGVP ++ D G+ F + V + + G+ +
Sbjct: 398 KWVEAIASPTNDATVVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKV 577
Query: 1225 ASVEHPQTNGQAES 1238
A+ HPQ +++S
Sbjct: 578 ATAYHPQKAERSKS 619
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 107 bits (266), Expect = 4e-23
Identities = 66/187 (35%), Positives = 98/187 (52%), Gaps = 6/187 (3%)
Frame = -2
Query: 837 EGEKTQGEWVLSVDGSSNNTGSGAGITIESPDKMIIEQSLKFEFKASNNQSEYEALIAGL 896
EG +W L DG+ N G G G I SP I + + F+ +NN +EYEA I G+
Sbjct: 558 EGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNNMAEYEACIFGI 379
Query: 897 RLAIELGVQKLFIKGDSQLVVKQVKGEYQVKDPQLSKYLEVVRRLMMEVKEIKIEHVPRG 956
AI++ ++ L I GDS LV+ Q+KGE++ L Y + RRL+ ++++ H+PR
Sbjct: 378 EEAIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYFTKVELHHIPRD 199
Query: 957 QNERADVLAKLASTGRLGNYQTV---IQETLPRPSIDLVEIKLKVVKSVNEGEL---PWM 1010
+N+ AD LA L+S R+ ++ V + L RPS V V+ E + PW
Sbjct: 198 ENQMADALATLSSMFRVNHWNDVPIIKVQRLERPS--HVFAIGNVIDQTGENVVDYKPWY 25
Query: 1011 ESIKTFL 1017
IK FL
Sbjct: 24 YDIKQFL 4
>BG587127 weakly similar to PIR|H84506|H84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(13%)
Length = 415
Score = 97.1 bits (240), Expect = 4e-20
Identities = 48/110 (43%), Positives = 72/110 (64%)
Frame = +3
Query: 861 GITIESPDKMIIEQSLKFEFKASNNQSEYEALIAGLRLAIELGVQKLFIKGDSQLVVKQV 920
GI + SP I++QS + EF ASNN++ YEALIAG+RLA L ++ + DSQLV Q
Sbjct: 3 GIRLTSPTNEILKQSFRLEFHASNNETNYEALIAGVRLAHGLKIRNIHAYCDSQLVASQF 182
Query: 921 KGEYQVKDPQLSKYLEVVRRLMMEVKEIKIEHVPRGQNERADVLAKLAST 970
GEY+ +D + YL++V++L ++ + +PR +N +AD LA LAS+
Sbjct: 183 SGEYEARDELMDTYLKLVQKLAQKLDYFALTRIPRSENVQADALAALASS 332
>TC83528
Length = 555
Score = 92.0 bits (227), Expect = 1e-18
Identities = 49/125 (39%), Positives = 79/125 (63%), Gaps = 4/125 (3%)
Frame = +3
Query: 173 PIVVTIRVNN-YVTKKVFLDQGSSADIIYGDAFDRLGLKESNLKPYKGTLVGFTGDRVSV 231
P+V+ +++NN + +VF++ S DI+Y AF ++ L+ES LKP +G L G G + V
Sbjct: 180 PMVIKLQINNNFSVLRVFVNPMSKVDILYWSAFLKMKLQESMLKPCQGFLKGTFGKGLPV 359
Query: 232 RGYVEIPTAFGEGEFVKKFQVKYLVLACRAN---YNVLLGRDTLNKVCAVISTAHLTVKY 288
+GY+++ T FG+GE K +V+Y V+ + YNV+LG +L + AV+S A T+KY
Sbjct: 360 KGYIDLDTTFGKGENTKTIKVRYFVVESPPSVSIYNVVLGWPSLKDLKAVLSVAEFTIKY 539
Query: 289 PACNG 293
P +G
Sbjct: 540 PVGDG 554
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 81.6 bits (200), Expect = 2e-15
Identities = 43/134 (32%), Positives = 73/134 (54%)
Frame = -1
Query: 333 QEGFSLDPRDEADDFRPQPLEETKQVQVKDKFLKIGSGLTTEQEDRLITLLGDNLDLFAW 392
QE ++ P E + EE K+ +K+G+ L + ++ LL + LD+FA
Sbjct: 384 QERKAIQPHQEEIELINLGTEENKRE------IKVGAALEEGVKRKIFQLLREYLDIFAC 223
Query: 393 TINDVPGIDPKVITHKLAIRPGATPVIQPMRRMSEEKHKAVQLETEKLIKARFIREVQYP 452
+ D+PG+DPK++ H++ +P PV +RR + ++ E +K I A F+ V+YP
Sbjct: 222 SYEDMPGLDPKIVEHRIPTKPECPPVR*KLRRTHPDMALKIKSEVQKQIDAGFLMTVEYP 43
Query: 453 TWLANVVMVKKANG 466
W+AN+V V K +G
Sbjct: 42 EWVANIVPVPKKDG 1
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 76.6 bits (187), Expect = 6e-14
Identities = 57/228 (25%), Positives = 103/228 (45%), Gaps = 6/228 (2%)
Frame = +2
Query: 1066 IMAEVHEGVCSSHIGGRSLAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEE 1125
++ E H+ + H GR+ ++++ F+WP + +V+ C+ C +A
Sbjct: 179 LVQESHDSTAAGH-PGRNGTLEIVSRKFFWPGQSQTVRRFVRNCDVCGGIHIWRQAKRGF 355
Query: 1126 LTTMMAPWPF-AMWGTDILGPFPVAKAQ-MKYIIVAVDYFTKWIEAEAVATITA-AKVRN 1182
L + P + D + P + + +Y+ V VD +K + E + T+ A A +
Sbjct: 356 LKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSKSVTLEEMDTMEAEACAQR 535
Query: 1183 FLWQRIVCRF---GVPMALVMDNGTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESA 1239
FL C + G+P ++V D G+ + REFC G+ ++ HPQT+G E
Sbjct: 536 FL----SCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGTERW 703
Query: 1240 NKVILKGLKKKLDEAKGLWAEELPGVLWAYNTTEQSSTKETPYRLTYG 1287
N+ I L+ + ++ W + LP V A SS TP+ + +G
Sbjct: 704 NQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHG 847
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 65.5 bits (158), Expect = 1e-10
Identities = 46/167 (27%), Positives = 72/167 (42%), Gaps = 29/167 (17%)
Frame = -2
Query: 759 KAALAILKTARRLRPYFQSFQVKVKTDV-PLRQVLQKPDLSGRLVSWSVELSEYDIQYEP 817
K A+ A+RLR Y + + + + P++ + +KP L+GR+ W + LSEYDI+Y
Sbjct: 635 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS 456
Query: 818 RGQVTVQSLIDFVA----------------------------ELTPTEGEKTQGEWVLSV 849
+ + L D +A E EG W L
Sbjct: 455 QKAIKGSILADHLAHQPLEDYRPIKFDFPDEEIMYLKMKDCDEPLFGEGPDPDSVWGLIF 276
Query: 850 DGSSNNTGSGAGITIESPDKMIIEQSLKFEFKASNNQSEYEALIAGL 896
DG+ N G+G G + +P I + + F +NN +EYEA I G+
Sbjct: 275 DGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGI 135
>TC92306
Length = 521
Score = 53.1 bits (126), Expect = 7e-07
Identities = 28/85 (32%), Positives = 38/85 (43%)
Frame = +2
Query: 893 IAGLRLAIELGVQKLFIKGDSQLVVKQVKGEYQVKDPQLSKYLEVVRRLMMEVKEIKIEH 952
I GL A G + + ++GD Q V KQ G ++V +P L L K + +EH
Sbjct: 2 ILGLNEARNQGYEHVHVRGDFQXVCKQFXGSWKVNNPNLRNLCNXAVELKSNFKSVSVEH 181
Query: 953 VPRGQNERADVLAKLASTGRLGNYQ 977
VPRG N AD A G +
Sbjct: 182 VPRGXNXAADAQANRGKNXGAGEVE 256
>TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNase H
{Arabidopsis thaliana}, partial (21%)
Length = 1071
Score = 51.6 bits (122), Expect = 2e-06
Identities = 33/108 (30%), Positives = 54/108 (49%), Gaps = 3/108 (2%)
Frame = +2
Query: 854 NNTGSGAGITIESPDKMII---EQSLKFEFKASNNQSEYEALIAGLRLAIELGVQKLFIK 910
N +GAG + + D ++ Q L + K S +EY AL+ GL+ A G + + K
Sbjct: 587 NRGPAGAGALLLAEDGSLLYGFRQGLGHQTKES---AEYRALLLGLKHASMKGFKYVTAK 757
Query: 911 GDSQLVVKQVKGEYQVKDPQLSKYLEVVRRLMMEVKEIKIEHVPRGQN 958
GDS+LV+ Q+ +++KD L K L +I+H+ R +N
Sbjct: 758 GDSELVINQILDPWKIKDEHLKKLCAEALELSDNFHSFRIQHISRERN 901
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 47.8 bits (112), Expect = 3e-05
Identities = 20/41 (48%), Positives = 27/41 (65%)
Frame = -1
Query: 456 ANVVMVKKANGKWRMCTDYTSLNKVCPKDSYPLPNVDKLVD 496
A ++ V+K +G +RMC DY NKV K+ YPLP +D L D
Sbjct: 238 AALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFD 116
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 40.0 bits (92), Expect = 0.006
Identities = 22/82 (26%), Positives = 39/82 (46%)
Frame = +2
Query: 1219 GIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEAKGLWAEELPGVLWAYNTTEQSSTK 1278
G + +S HP ++GQ+E+ NK L+ + W++ P + YNT+ S
Sbjct: 65 GTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEYWYNTSYNISAA 244
Query: 1279 ETPYRLTYGTDAMLSVEIENQS 1300
TP++ YG D + + + S
Sbjct: 245 MTPFKALYGRDLSMLIRSKGSS 310
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 38.5 bits (88), Expect = 0.019
Identities = 31/115 (26%), Positives = 55/115 (46%), Gaps = 4/115 (3%)
Frame = +2
Query: 703 PVLSKPTPGVPLVLYLAVTDKAVS---TVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEK 759
P+L P L+ Y+ TD +++ VL Q E KVI + S L+ E Y +
Sbjct: 17 PILVLPE----LITYVVYTDASITGLGCVLTQHE----KVIAYASRQLRKHEGNYPTHDL 172
Query: 760 AALAILKTARRLRPYFQSFQVKVKTD-VPLRQVLQKPDLSGRLVSWSVELSEYDI 813
A++ + R Y +V++ TD L+ + +P+L+ R W +++YD+
Sbjct: 173 EMAAVVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDL 337
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 37.4 bits (85), Expect = 0.042
Identities = 23/68 (33%), Positives = 31/68 (44%), Gaps = 5/68 (7%)
Frame = +1
Query: 1198 LVMDNGTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEA--- 1254
L+ DN +F SS EFC GI +PQ NG AE + +L+ + L A
Sbjct: 295 LITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL* 474
Query: 1255 --KGLWAE 1260
+ LW E
Sbjct: 475 N*RDLWVE 498
>BG448304
Length = 637
Score = 37.4 bits (85), Expect = 0.042
Identities = 20/59 (33%), Positives = 31/59 (51%)
Frame = +2
Query: 46 RWCEYHKALGHTTDNCWNLRREIDRLIKAGHLANFVKDTATPEVAKITQEDKGKGKEIV 104
++C +HK H TD+C +L+ I+ LI+ G L K+ PE + K K+IV
Sbjct: 107 KYCRFHKCHRHLTDDCIHLKDTIEILIQRGRLNQLTKN-PEPEKQTVKLITDKKNKDIV 280
>TC77786
Length = 1094
Score = 37.0 bits (84), Expect = 0.054
Identities = 15/34 (44%), Positives = 21/34 (61%)
Frame = +3
Query: 48 CEYHKALGHTTDNCWNLRREIDRLIKAGHLANFV 81
C YH++ GH T+ C+ LR I+ LIK L F+
Sbjct: 825 CCYHRSRGHDTEECFQLREAIEDLIKKVKLGRFI 926
>TC81434
Length = 363
Score = 28.1 bits (61), Expect(2) = 0.40
Identities = 13/24 (54%), Positives = 15/24 (62%)
Frame = +1
Query: 1390 AYKLEQLSGQKVPRTWNASHLKQY 1413
AYKLE + R WNA+HLK Y
Sbjct: 154 AYKLEGRARNIYLRLWNATHLKLY 225
Score = 24.6 bits (52), Expect(2) = 0.40
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +2
Query: 1368 KHNKLSPNWGGPY 1380
KH KL+ +W GPY
Sbjct: 89 KHGKLAASWKGPY 127
>TC78806 similar to GP|8978273|dbj|BAA98164.1 receptor protein kinase-like
{Arabidopsis thaliana}, partial (28%)
Length = 1962
Score = 33.9 bits (76), Expect = 0.46
Identities = 58/257 (22%), Positives = 109/257 (41%), Gaps = 17/257 (6%)
Frame = +2
Query: 780 VKVKTDVPLRQVLQKPDLSGRLVSWSVELSEYDIQYE--PRGQVTVQSLIDFVAELTPTE 837
V+ KT++ L + +L LV + E E + YE P G +L+D ++
Sbjct: 692 VEFKTEIELLSRVHHKNLVS-LVGFCYEKGEQMLVYEYVPNG-----TLLDSLS------ 835
Query: 838 GEKTQGEWVLSVDGSSNNTGSGAGITI--ESPDKMIIEQSLKFEFKASNNQSEYEALIAG 895
+ G W+ + G+ G+T E D II + +K S+N LIA
Sbjct: 836 --RKSGIWMDWIRRLKVTLGAARGLTYLHELADPPIIHRDIK-----SSNILLDNHLIAK 994
Query: 896 LRLAIELGVQKLFIKGDSQLVVKQVKGEYQVKDPQLSKYLEVVRR--------LMMEVKE 947
+ + G+ KL + + V QVKG DP+ ++ + LM+E+
Sbjct: 995 VA---DFGLSKLLVDSERGHVTTQVKGTMGYLDPEYYMTQQLTEKSDVYSFGVLMLELAT 1165
Query: 948 IKIEHVPRGQNERADVLAKLASTGRLGNYQTVIQETL-----PRPSIDLVEIKLKVVKSV 1002
+ + + +G+ +V+ + ++ L N +++ ++L P+ VE+ L+ VK
Sbjct: 1166 SR-KPIEQGKYIVREVMRVMDTSKELYNLHSILDQSLLKGTRPKGLERYVELALRCVKEY 1342
Query: 1003 NEGELPWMESIKTFLEN 1019
E P M + +E+
Sbjct: 1343 -AAERPSMAEVAKEIES 1390
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,610,734
Number of Sequences: 36976
Number of extensions: 493294
Number of successful extensions: 2340
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 2307
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2334
length of query: 1414
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1306
effective length of database: 5,021,319
effective search space: 6557842614
effective search space used: 6557842614
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0114.12


